He, Miao, et al. "Cell-type-based analysis of microRNA profiles in the mouse brain." Neuron 73.1 (2012): 35-48.
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6985 | 1.32211912307e-07 | 0.724203377061 | 0.940277304239 | Down in P28-KO |
| Mir1982 | 8.551190116e-07 | 0.999048950327 | 0.417918028877 | Up in P28-KO |
| Mir8091 | 8.6737612992e-06 | 4.29418330277e-07 | 2.02268609066 | Down in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir412 | 5.50250529252e-07 | 0.420462739775 | 1.4032394844 | Down in P28R-WT |
| Mir7213 | 6.05856406335e-05 | 0.763796659629 | 0.803514321589 | Down in P28R-WT |
| Mir7664 | 0.000260515021712 | 0.985677562593 | 0.372750140792 | Down in P28R-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6954 | 2.32011054369e-07 | 0.485922957335 | 1.03213082754 | Down in P28-WT |
| Mir409 | 3.16798732247e-07 | 1.0 | 0.0 | Down in P28-WT |
| Mir6915 | 4.43301199637e-07 | 0.852687000215 | 0.715736071976 | Down in P28-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6966 | 4.88659584019e-07 | 1.0 | 0.0 | Down in P10-KO |
| Mir324 | 7.01447213713e-06 | 0.942325355515 | 0.433263946166 | Down in P10-KO |
| Mir212 | 2.00615028338e-05 | 0.944120205154 | 0.546755220724 | Down in P10-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir7052 | 3.89303944637e-09 | 0.842594085013 | 0.935966747359 | Up in P28-KO |
| Mir9768 | 9.49971670217e-08 | 0.942284675862 | 0.903000298189 | Down in P28-KO |
| Mir7240 | 9.79275536217e-08 | 0.962941193092 | 0.864954840315 | Up in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 6.18838313926e-13 | 1.76117361048e-90 | 2.20631862566 | Up in P28-WT |
| Mir7052 | 6.81166234529e-12 | 0.99815374504 | 0.831511445591 | Up in P28-WT |
| Mir212 | 1.11044506923e-11 | 1.71434096e-88 | 2.20202339 | Up in P28-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir374b | 6.37208415899e-06 | 1.0 | nan | Up in P28L-KO |
| Mir132 | 2.26206794822e-05 | 3.15157607324e-67 | 3.02454303755 | Down in P28L-KO |
| Mir6909 | 9.17317672554e-05 | 0.000260127785293 | 1.30441294717 | Up in P28L-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 2.21535609917e-05 | 1.82973026956e-27 | 2.9016759516 | Down in P28-KO |
| Mir212 | 0.000151462987987 | 2.23594106337e-25 | 2.8148654465 | Down in P28-KO |
| Mir7052 | 0.00126228513944 | 0.979833037891 | 0.698124709032 | Down in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6990 | 0.00311252911028 | 1.0 | nan | Down in P28L-KO |
| Mir7578 | 0.00433541734335 | 1.0 | nan | Up in P28L-KO |
| Mir7004 | 0.0112640733128 | 1.0 | nan | Down in P28L-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir7090 | 3.575379944e-05 | 0.793175063285 | 0.837139609076 | Down in P28R-KO |
| Mir5119 | 0.000299174124507 | 0.416492543237 | 1.41545304848 | Down in P28R-KO |
| Mir700 | 0.0004381964198 | 0.879497198443 | 0.592501969397 | Down in P28R-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6985 | 1.32211912307e-07 | 0.950474439465 | 0.758010948487 | Down in P28-KO |
| Mir1982 | 8.551190116e-07 | 1.0 | 0.0 | Up in P28-KO |
| Mir8091 | 8.6737612992e-06 | 1.19922405369e-05 | 2.34486849351 | Down in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir412 | 5.50250529252e-07 | 1.0 | 0.0 | Down in P28R-WT |
| Mir7213 | 6.05856406335e-05 | 0.875934818389 | 0.478748951191 | Down in P28R-WT |
| Mir7664 | 0.000260515021712 | 0.848710165285 | 0.530335566168 | Down in P28R-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6954 | 2.32011054369e-07 | 0.889079517354 | 0.453566142207 | Down in P28-WT |
| Mir409 | 3.16798732247e-07 | 1.0 | 0.0 | Down in P28-WT |
| Mir6915 | 4.43301199637e-07 | 1.0 | 0.0 | Down in P28-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6966 | 4.88659584019e-07 | 1.0 | nan | Down in P10-KO |
| Mir324 | 7.01447213713e-06 | 1.0 | nan | Down in P10-KO |
| Mir212 | 2.00615028338e-05 | 1.0 | nan | Down in P10-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir7052 | 3.89303944637e-09 | 0.585646291958 | 0.985355503644 | Up in P28-KO |
| Mir9768 | 9.49971670217e-08 | 0.862175712974 | 0.923666076378 | Down in P28-KO |
| Mir7240 | 9.79275536217e-08 | 0.626784410251 | 0.973048892017 | Up in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 6.18838313926e-13 | 2.30283938795e-91 | 2.38847225751 | Up in P28-WT |
| Mir7052 | 6.81166234529e-12 | 0.998368221695 | 0.810687691148 | Up in P28-WT |
| Mir212 | 1.11044506923e-11 | 2.57082325135e-89 | 2.38256627949 | Up in P28-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir374b | 6.37208415899e-06 | 1.0 | nan | Up in P28L-KO |
| Mir132 | 2.26206794822e-05 | 2.50215945276e-24 | 3.14909686894 | Down in P28L-KO |
| Mir6909 | 9.17317672554e-05 | 0.0141146214793 | 1.28462185418 | Up in P28L-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 2.21535609917e-05 | 6.62433960297e-06 | 2.8721240633 | Down in P28-KO |
| Mir212 | 0.000151462987987 | 1.27040983602e-05 | 2.80967859309 | Down in P28-KO |
| Mir7052 | 0.00126228513944 | 0.971197072246 | 0.420187912646 | Down in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6990 | 0.00311252911028 | 1.0 | nan | Down in P28L-KO |
| Mir7578 | 0.00433541734335 | 1.0 | nan | Up in P28L-KO |
| Mir7004 | 0.0112640733128 | 1.0 | nan | Down in P28L-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir7090 | 3.575379944e-05 | 0.368454503581 | 1.19331836908 | Down in P28R-KO |
| Mir5119 | 0.000299174124507 | 1.0 | 0.0 | Down in P28R-KO |
| Mir700 | 0.0004381964198 | 0.819356426164 | 0.585320127343 | Down in P28R-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6985 | 1.32211912307e-07 | 0.996442649284 | 0.376218669286 | Down in P28-KO |
| Mir1982 | 8.551190116e-07 | 1.0 | 0.0 | Up in P28-KO |
| Mir8091 | 8.6737612992e-06 | 0.000110217903784 | 3.31895860077 | Down in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir412 | 5.50250529252e-07 | 1.0 | 0.0 | Down in P28R-WT |
| Mir7213 | 6.05856406335e-05 | 1.0 | 0.0 | Down in P28R-WT |
| Mir7664 | 0.000260515021712 | 1.0 | 0.0 | Down in P28R-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6954 | 2.32011054369e-07 | 1.0 | 0.0 | Down in P28-WT |
| Mir409 | 3.16798732247e-07 | 1.0 | 0.0 | Down in P28-WT |
| Mir6915 | 4.43301199637e-07 | 1.0 | 0.0 | Down in P28-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6966 | 4.88659584019e-07 | 1.0 | nan | Down in P10-KO |
| Mir324 | 7.01447213713e-06 | 1.0 | nan | Down in P10-KO |
| Mir212 | 2.00615028338e-05 | 1.0 | nan | Down in P10-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir7052 | 3.89303944637e-09 | 0.790856355384 | 0.924413407821 | Up in P28-KO |
| Mir9768 | 9.49971670217e-08 | 0.788195033165 | 0.934231327095 | Down in P28-KO |
| Mir7240 | 9.79275536217e-08 | 0.793670811275 | 0.909586905086 | Up in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 6.18838313926e-13 | 8.84363128396e-81 | 2.530731629 | Up in P28-WT |
| Mir7052 | 6.81166234529e-12 | 0.985700947181 | 0.837330985345 | Up in P28-WT |
| Mir212 | 1.11044506923e-11 | 2.39882769045e-82 | 2.56667890875 | Up in P28-WT |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir374b | 6.37208415899e-06 | 1.0 | nan | Up in P28L-KO |
| Mir132 | 2.26206794822e-05 | 0.00719604753125 | 2.3293916541 | Down in P28L-KO |
| Mir6909 | 9.17317672554e-05 | 0.0414288055279 | 1.64926712008 | Up in P28L-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 2.21535609917e-05 | 0.0833942614805 | 3.56539538892 | Down in P28-KO |
| Mir212 | 0.000151462987987 | 0.0790191857883 | 3.65544918999 | Down in P28-KO |
| Mir7052 | 0.00126228513944 | 1.0 | 0.0 | Down in P28-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir6990 | 0.00311252911028 | 1.0 | nan | Down in P28L-KO |
| Mir7578 | 0.00433541734335 | 1.0 | nan | Up in P28L-KO |
| Mir7004 | 0.0112640733128 | 1.0 | nan | Down in P28L-KO |
| MicroRNA | CyberT P-Value | Enriched P-Value | OddsRatio | Direction |
| Mir7090 | 3.575379944e-05 | 0.200945034471 | 1.80752635316 | Down in P28R-KO |
| Mir5119 | 0.000299174124507 | 1.0 | 0.0 | Down in P28R-KO |
| Mir700 | 0.0004381964198 | 1.0 | 0.0 | Down in P28R-KO |
| GO Biological Process | Pos/Neg Enrichment | Fold | P-Value |
| Palate development | + | > 5 | 1.24e-03 |
| Cell cycle arrest | + | > 5 | 2.51e-02 |
| Negative regulation of transcription from rna polymerase ii promoter | + | > 5 | 5.70e-23 |
| Negative regulation of transcription, dna-templated | + | > 5 | 1.96e-30 |
| Histone modification | + | > 5 | 6.68e-06 |
| Negative regulation of nucleic acid-templated transcription | + | > 5 | 3.69e-30 |
| Covalent chromatin modification | + | > 5 | 7.54e-06 |
| Negative regulation of rna biosynthetic process | + | > 5 | 6.33e-30 |
| Chromatin modification | + | > 5 | 2.57e-10 |
| Negative regulation of rna metabolic process | + | > 5 | 3.07e-29 |
| Transcription, dna-templated | + | > 5 | 1.64e-61 |
| Nucleic acid-templated transcription | + | > 5 | 1.71e-61 |
| Rna biosynthetic process | + | > 5 | 2.27e-61 |
| Negative regulation of cellular macromolecule biosynthetic process | + | > 5 | 1.82e-29 |
| Chromatin organization | + | > 5 | 4.63e-11 |
| Negative regulation of macromolecule biosynthetic process | + | > 5 | 1.44e-28 |
| Negative regulation of nucleobase-containing compound metabolic process | + | > 5 | 2.01e-27 |
| Negative regulation of nitrogen compound metabolic process | + | > 5 | 4.55e-27 |
| Negative regulation of cellular biosynthetic process | + | > 5 | 8.61e-28 |
| Negative regulation of gene expression | + | > 5 | 1.78e-28 |
| Rhythmic process | + | > 5 | 1.24e-02 |
| Nucleobase-containing compound biosynthetic process | + | > 5 | 1.11e-57 |
| Negative regulation of biosynthetic process | + | > 5 | 1.91e-27 |
| Heterocycle biosynthetic process | + | > 5 | 1.01e-56 |
| Transcription from rna polymerase ii promoter | + | > 5 | 2.55e-13 |
| Aromatic compound biosynthetic process | + | > 5 | 1.34e-56 |
| Cellular nitrogen compound biosynthetic process | + | > 5 | 1.31e-55 |
| Regulation of transcription, dna-templated | + | > 5 | 8.15e-82 |
| Positive regulation of rna biosynthetic process | + | > 5 | 2.56e-26 |
| Regulation of transcription from rna polymerase ii promoter | + | > 5 | 6.51e-34 |
| Regulation of nucleic acid-templated transcription | + | > 5 | 1.28e-81 |
| Regulation of rna biosynthetic process | + | > 5 | 1.57e-81 |
| Organic cyclic compound biosynthetic process | + | > 5 | 4.11e-55 |
| Positive regulation of rna metabolic process | + | > 5 | 7.02e-27 |
| Response to growth factor | + | > 5 | 2.51e-02 |
| Positive regulation of nucleic acid-templated transcription | + | > 5 | 3.10e-25 |
| Positive regulation of transcription, dna-templated | + | > 5 | 3.10e-25 |
| Regulation of rna metabolic process | + | > 5 | 3.01e-80 |
| Positive regulation of nucleobase-containing compound metabolic process | + | > 5 | 6.11e-29 |
| Positive regulation of nitrogen compound metabolic process | + | > 5 | 1.52e-28 |
| Positive regulation of macromolecule biosynthetic process | + | > 5 | 2.36e-26 |
| Regulation of cellular macromolecule biosynthetic process | + | > 5 | 8.55e-78 |
| Positive regulation of transcription from rna polymerase ii promoter | + | > 5 | 3.37e-14 |
| Chromosome organization | + | > 5 | 4.62e-10 |
| Rna metabolic process | + | > 5 | 1.22e-52 |
| Cellular macromolecule biosynthetic process | + | > 5 | 1.55e-52 |
| Positive regulation of gene expression | + | > 5 | 1.57e-25 |
| Regulation of macromolecule biosynthetic process | + | > 5 | 1.59e-76 |
| Macromolecule biosynthetic process | + | > 5 | 3.07e-52 |
| Regulation of nucleobase-containing compound metabolic process | + | > 5 | 3.41e-76 |
| Regulation of nitrogen compound metabolic process | + | > 5 | 2.20e-75 |
| Positive regulation of cellular biosynthetic process | + | > 5 | 9.12e-25 |
| Positive regulation of biosynthetic process | + | > 5 | 2.17e-24 |
| Regulation of cellular biosynthetic process | + | > 5 | 3.11e-74 |
| Regulation of gene expression | + | > 5 | 5.40e-74 |
| Regulation of biosynthetic process | + | > 5 | 1.21e-73 |
| Gene expression | + | > 5 | 1.94e-50 |
| Nucleic acid metabolic process | + | > 5 | 5.75e-49 |
| In utero embryonic development | + | > 5 | 1.90e-02 |
| Negative regulation of macromolecule metabolic process | + | > 5 | 1.21e-24 |
| Cellular response to oxygen-containing compound | + | > 5 | 1.09e-03 |
| Negative regulation of cellular metabolic process | + | > 5 | 6.50e-24 |
| Skeletal system development | + | > 5 | 2.98e-02 |
| Cellular biosynthetic process | + | > 5 | 3.91e-46 |
| Organic substance biosynthetic process | + | > 5 | 4.04e-45 |
| Nucleobase-containing compound metabolic process | + | > 5 | 7.36e-45 |
| Negative regulation of metabolic process | + | > 5 | 7.51e-23 |
| Biosynthetic process | + | > 5 | 2.26e-44 |
| Heterocycle metabolic process | + | > 5 | 1.58e-43 |
| Cellular aromatic compound metabolic process | + | > 5 | 3.34e-43 |
| Cellular nitrogen compound metabolic process | + | 4.85 | 7.94e-42 |
| Positive regulation of macromolecule metabolic process | + | 4.83 | 1.23e-21 |
| Organic cyclic compound metabolic process | + | 4.80 | 1.83e-41 |
| Chordate embryonic development | + | 4.74 | 1.15e-02 |
| Positive regulation of cellular metabolic process | + | 4.70 | 3.37e-22 |
| Embryo development ending in birth or egg hatching | + | 4.69 | 1.30e-02 |
| Negative regulation of cell death | + | 4.62 | 4.48e-04 |
| Nitrogen compound metabolic process | + | 4.62 | 6.18e-42 |
| Regulation of macromolecule metabolic process | + | 4.61 | 6.37e-60 |
| Regulation of primary metabolic process | + | 4.60 | 6.92e-60 |
| Apoptotic process | + | 4.55 | 1.34e-03 |
| Cellular response to endogenous stimulus | + | 4.55 | 4.35e-02 |
| Programmed cell death | + | 4.43 | 1.97e-03 |
| Regulation of cellular metabolic process | + | 4.40 | 5.62e-58 |
| Negative regulation of apoptotic process | + | 4.29 | 1.59e-02 |
| Circulatory system development | + | 4.26 | 3.86e-02 |
| Cardiovascular system development | + | 4.26 | 3.86e-02 |
| Cell death | + | 4.25 | 3.55e-03 |
| Negative regulation of programmed cell death | + | 4.22 | 1.95e-02 |
| Death | + | 4.22 | 3.90e-03 |
| Organ morphogenesis | + | 4.20 | 9.09e-03 |
| Positive regulation of metabolic process | + | 4.07 | 1.22e-19 |
| Epithelium development | + | 3.97 | 1.91e-02 |
| Negative regulation of signal transduction | + | 3.95 | 2.02e-02 |
| Regulation of metabolic process | + | 3.92 | 4.82e-53 |
| Cellular macromolecule metabolic process | + | 3.90 | 1.21e-43 |
| Embryo development | + | 3.83 | 1.44e-02 |
| Cellular response to stress | + | 3.82 | 7.73e-04 |
| Macromolecular complex subunit organization | + | 3.78 | 1.85e-06 |
| Regulation of apoptotic process | + | 3.76 | 4.69e-04 |
| Regulation of cell death | + | 3.75 | 1.07e-04 |
| Negative regulation of cellular process | + | 3.74 | 7.06e-21 |
| Regulation of programmed cell death | + | 3.71 | 5.81e-04 |
| Cellular response to organic substance | + | 3.57 | 9.31e-03 |
| Tissue development | + | 3.52 | 7.20e-04 |
| Negative regulation of biological process | + | 3.50 | 8.62e-20 |
| Macromolecule metabolic process | + | 3.49 | 2.00e-39 |
| Regulation of cell proliferation | + | 3.44 | 4.30e-03 |
| Regulation of multicellular organismal development | + | 3.36 | 8.50e-04 |
| Positive regulation of cellular process | + | 3.33 | 1.43e-18 |
| Cellular response to chemical stimulus | + | 3.29 | 2.41e-03 |
| Response to external stimulus | + | 3.19 | 4.80e-02 |
| Regulation of cell differentiation | + | 3.15 | 1.74e-02 |
| Cellular protein modification process | + | 3.14 | 1.29e-04 |
| Protein modification process | + | 3.14 | 1.29e-04 |
| Macromolecule modification | + | 3.07 | 1.07e-04 |
| Positive regulation of biological process | + | 3.00 | 1.60e-16 |
| Cellular metabolic process | + | 2.99 | 1.78e-33 |
| Primary metabolic process | + | 2.89 | 3.43e-32 |
| Regulation of developmental process | + | 2.86 | 2.97e-03 |
| Regulation of signal transduction | + | 2.81 | 1.35e-03 |
| Organic substance metabolic process | + | 2.78 | 1.07e-30 |
| Organelle organization | + | 2.72 | 1.54e-03 |
| Organ development | + | 2.66 | 2.67e-03 |
| Regulation of cell communication | + | 2.64 | 1.75e-03 |
| Response to stress | + | 2.63 | 5.71e-03 |
| Regulation of multicellular organismal process | + | 2.61 | 1.03e-02 |
| Metabolic process | + | 2.57 | 1.20e-27 |
| Regulation of signaling | + | 2.57 | 5.51e-03 |
| Cellular protein metabolic process | + | 2.56 | 5.93e-03 |
| Cellular component organization | + | 2.36 | 6.59e-05 |
| System development | + | 2.31 | 3.66e-03 |
| Cell differentiation | + | 2.30 | 2.39e-02 |
| Cellular component organization or biogenesis | + | 2.28 | 1.86e-04 |
| Regulation of response to stimulus | + | 2.28 | 4.17e-02 |
| Regulation of cellular process | + | 2.25 | 2.41e-29 |
| Multicellular organismal development | + | 2.22 | 1.69e-03 |
| Single-organism developmental process | + | 2.21 | 1.28e-04 |
| Developmental process | + | 2.19 | 1.52e-04 |
| Regulation of biological process | + | 2.16 | 1.25e-27 |
| Anatomical structure development | + | 2.14 | 4.49e-03 |
| Biological regulation | + | 2.07 | 8.21e-26 |
| Cellular process | + | 1.58 | 7.09e-11 |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.999998593648 | 0.379307274508 | Up in P28R-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.999999999994 | 0.282341505085 | Up in P28-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.997817145581 | 0.316889621741 | Down in P10-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 8.0282762784e-26 | 1.52378835118 | Up in P28-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 2.00557975416e-31 | 2.1052498853 | Down in P28L-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 2.43568184168e-13 | 2.03248610855 | Down in P28-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 3.48726808465e-13 | 2.45299264888 | Down in P28L-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.998328515297 | 0.219598948399 | Up in P28R-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.999999437104 | 0.252871516339 | Up in P28-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | nan | Down in P10-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.12324909962e-30 | 1.65278481253 | Up in P28-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.26818145025e-12 | 2.20176840369 | Down in P28L-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.00130283253386 | 2.06164659791 | Down in P28-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.000939708451145 | 2.58506153388 | Down in P28L-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | nan | Up in P28R-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.999880442366 | 0.10430950049 | Up in P28-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | nan | Down in P10-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 2.36569994701e-31 | 1.77226972721 | Up in P28-WT |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.0513863414703 | 1.75239960823 | Down in P28L-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.149223920241 | 2.68224429831 | Down in P28-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 0.447367049331 | 2.08619000979 | Down in P28L-KO |
| MicroRNA | Enriched P-Value | OddsRatio | Direction |
| Mir132 | 1.0 | NA | NA |
| Cd24a | Lipg | Tac1 | Egr4 | Actr3b | Nudt10 | Fbxo9 | Yaf2 |
| Tsnax | Frat1 | Gnb4 | Cyp51 | Ctgf | Sst | Phlda3 | Mrpl36 |
| Bik | Ier5 | Twist2 | Gpr133 | Rprm | Cdkn1a | Kcnk2 | Pdp1 |
| Cinp | Fst | Kcnv1 | F8a | Bcl7c |
| Pcdha2 | Clmn | Med12 | Pxn | Cc2d1b | Col24a1 | 1700109k24rik | Gigyf1 |
| Fn1 | Zfp652 | Mllt6 | H2-t24 | Spen | Zfp455 | A330023f24rik | Hmbox1 |
| Sfi1 | Cntnap1 | Spp1 | Gm14137 | Mmp13 | E230029c05rik | Celsr3 | Podxl |
| Cdkn1a | Fst | Cd24a | Lipg |
| Gigyf1 | Fn1 | Sfi1 | Celsr3 | H2-t24 | Pxn | Cc2d1b | Col24a1 |
| None |
| H2-t24 |
| Snx11 | Fnbp1 | *Med22 | Rnps1 | Snx11 | *Sox13 | Gsta3 | *Yeats2 |
| Mical3 | Rnf135 | Ipo9 | Fbxo9 | Klc4 | Bicd2 | Adamts3 | Hkdc1 |
| Hnrnpm | Stx1a | Mkrn1 | Sept5 | Rimkla | Nit1 | Brap | Lysmd4 |
| Ubtf | Pamr1 | Frat1 | *Il1a | *Pbx4 | Repin1 | Papd7 |
| Bptf | Nfat5 | Xkr4 | Macf1 | Smg1 | Taf1d | Ktn1 | Hmbox1 |
| Med13l | A330023f24rik | Rab30 | Prpf4b | C130060c02rik | Nav1 | Ttc14 | A230046k03rik |
| Prpf38b | Lnpep | Zfc3h1 | Trpm3 |
| Ptgs2 | Egr4 | Bc022687 | Mrps12 | Fkbp2 | Cst3 | Tomm7 | Cmbl |
| Rpsa | Hn1 | Nnmt | Rpl36al | Rpl18 | Tob1 | Btg2 | Sst |
| Cort | Ldlr | Dnajc15 | Cdkn1a | Rpl13a | Grasp |
| Xkr4 | Taf1d | Smg1 | Prpf4b | Trpm3 |
| Snx11 | Brap | Adamts3 | Snx11 |
| Tada2b | Slc16a9 | E430025e21rik |
| Hn1 | Pcbp2 | Lgi4 | Akirin1 | Lipg | Arf3 | Acss2 | Arf1 |
| Rgs19 | Hn1 | Frat1 | Bc022687 | Gpr133 | Sept5 | Adamts15 |
| Usp38 | *Jmy | Arfgef2 | E430025e21rik | Fubp1 | *Elk4 | Ddx10 | Mdc1 |
| Prpf4b | Phf3 | Rbm26 | Dnajc13 | Arfgef2 | Prkx | Usp24 |
| Pgm5 | Ptpn21 | Snupn | Hspa12b | Gatsl3 | Arhgap27 | Ucp2 | Als2cl |
| Pwp1 |
| Crk | Tada2b | Plcxd2 | Rnf24 | *Pik3r1 | Fam65b | Fam49a | Hmgcll1 |
| Creg2 | Adamts2 | Rap1gds1 | Camsap1 | Fam73a | Golga3 | Lrfn5 | Ppih |
| Syn2 | Ppp2r5a | Mapre2 |
| No Targets Found |
| Plcxd2 |
| Ptgs2 | Cyp51 | Btg2 | Lipg | Scgb1c1 | Cdkn1a | Egr1 | Arl5b |
| Plcxd2 | Dnajb5 | Pdp1 | Tfrc | Tiparp | Egr4 | Homer1 | P4ha1 |
| Kcnv1 | Trib1 |
| Cygb | Amz1 | Zfp467 | 4930578m01rik | Mapk3 | Pik3ip1 | Fads6 |
| Ptgs2 | Egr1 | Cyp51 | Tiparp | Trib1 | Kcnv1 |
| None |
| Tnfsf13 | Vasn | Col1a2 | Gm10451 |
| 1700003m02rik | Pstpip1 | Gng11 |
| No Targets |
| No Targets |
| No Targets |
| No Targets |
| Ptgs2 | E330009j07rik | Lipg | Egr1 | Egr4 | Pggt1b | Pcdhac1 | Hkdc1 |
| Pde7b | Scn1a | Kcnv1 | Adra1a | Btg2 | Chsy3 | Trib1 | Homer1 |
| Ipcef1 | Mex3b | Amd1 | Rragd | Cdkn1a | Ai593442 | Pdp1 | Fst |
| 1700047m11rik | Cmbl | Ntng1 | Akap2 | Adamts2 | Klrg1 | Lmo2 | B430306n03rik |
| Id3 | Plac8l1 | Mapk3 | Bag3 | Mdga1 | Btc | Ikbke | Slc9a2 |
| Utp14a | Cd3e |
| Ptgs2 | Btg2 | Lipg | Egr1 | Egr4 | Trib1 | Kcnv1 |
| Cmbl | Ntng1 |
| Ptgs2 | Btg2 |
| No Targets |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Ribosomal protein | 6.354545999490477 | 2.151639158733559e-52 | 9.479542834426517e-51 | 3.415560744806351e-51 | 3.415560744806351e-51 | 6.543219817636688e-54 | 5.273625389052841e-55 |
| Ribonucleoprotein | 5.1631000578369 | 2.927390055218994e-52 | 1.7312670820520133e-50 | 3.1189520357233695e-51 | 6.237904071446739e-51 | 1.1950007799706397e-53 | 1.43499512510735e-54 |
| Go:0005739~mitochondrion | 2.3773348145049797 | 1.8734098522303243e-46 | 4.150580887026649e-45 | 1.3265583842524502e-45 | 1.3265583842524502e-45 | 2.921934767075882e-48 | 1.0644374160399571e-48 |
| Go:0005840~ribosome | 5.55374775583483 | 5.915305494507822e-47 | 4.7323522897151645e-45 | 7.56248556290047e-46 | 1.512497112580094e-45 | 3.331491437401088e-48 | 3.3609690309703535e-49 |
| Mitochondrion | 2.862466843501326 | 1.1302100112564573e-40 | 2.7697699758119257e-39 | 3.3265730801453678e-40 | 9.979719240436104e-40 | 1.911823609278947e-42 | 5.5402451532179275e-43 |
| Acetylation | 1.9452462850460623 | 6.3958725383610245e-40 | 9.675409141966452e-39 | 8.715332646429345e-40 | 3.486133058571738e-39 | 6.678415821018655e-42 | 3.1352316364514825e-42 |
| Kidney | 1.9758653150959822 | 1.6067633642441235e-35 | 3.171425224839525e-34 | 5.502020132649646e-35 | 5.502020132649646e-35 | 2.4896018699772155e-37 | 1.1559448663626788e-37 |
| Mmu03010:ribosome | 6.597664050995502 | 1.1166187683206011e-34 | 1.9855743313879283e-32 | 2.8612666976239504e-33 | 2.8612666976239504e-33 | 1.6165348574146612e-35 | 1.0059628543428839e-36 |
| Go:0030529~ribonucleoprotein complex | 3.2069761477301872 | 1.1127874557996016e-33 | 3.6623556638995424e-32 | 3.901725842155352e-33 | 1.1705177526466054e-32 | 2.578232935344946e-35 | 6.322655998861372e-36 |
| Mitochondrion inner membrane | 4.981657027728255 | 7.303256557844224e-33 | 4.046000585529822e-31 | 2.915617559786089e-32 | 1.4578087798930444e-31 | 2.792737126231886e-34 | 3.5800277244334433e-35 |
| Mmu00190:oxidative phosphorylation | 5.309294296367905 | 2.0772910273566638e-32 | 1.1193927480952249e-30 | 8.065377208638142e-32 | 1.6130754417276284e-31 | 9.113420574732363e-34 | 9.357166789894882e-35 |
| Go:0044429~mitochondrial part | 2.977469266928886 | 9.696239589918892e-32 | 2.881596665127319e-30 | 2.3024525483329068e-31 | 9.209810193331627e-31 | 2.0285925535972748e-33 | 5.509227039726643e-34 |
| Go:0005743~mitochondrial inner membrane | 3.7920326071133967 | 1.004767997082062e-31 | 4.209222922599209e-30 | 2.6906016825811747e-31 | 1.3453008412905875e-30 | 2.963217712093805e-33 | 5.708909074329898e-34 |
| Go:0006412~translation | 3.8236634905984253 | 4.9987741793308815e-31 | 6.96313926626644e-30 | 1.1885728062772585e-29 | 1.1885728062772585e-29 | 3.855247506575603e-33 | 7.573900271713457e-34 |
| Go:0019866~organelle inner membrane | 3.6695207844220414 | 5.234849689758962e-31 | 2.0884510074489314e-29 | 1.1124756869138406e-30 | 6.674854121483043e-30 | 1.4702321853486878e-32 | 2.974346414635774e-33 |
| Go:0005740~mitochondrial envelope | 3.2725920279906515 | 1.0993310688298755e-29 | 3.7204109720785356e-28 | 1.6986752671180653e-29 | 1.1890726869826458e-28 | 2.619102834763537e-31 | 6.246199254715202e-32 |
| Mmu05012:parkinson's disease | 5.034624262631444 | 6.949864234743548e-30 | 6.7400155331351745e-28 | 3.2375153260144287e-29 | 9.712545978043287e-29 | 5.487314111888863e-31 | 6.261138950219412e-32 |
| Go:0031966~mitochondrial membrane | 3.3246233705409414 | 5.74931324854718e-29 | 1.9891680761179886e-27 | 7.946922553989646e-29 | 6.357538043191717e-28 | 1.4003387760334178e-30 | 3.2666552548563523e-31 |
| Mmu05016:huntington's disease | 4.27826667059089 | 3.190355599119923e-29 | 2.3112623248456886e-27 | 8.326479919453902e-29 | 3.3305919677815607e-28 | 1.881690377277718e-30 | 2.874194233441372e-31 |
| Mammary gland | 1.8792152283699464 | 1.6829487830423295e-28 | 3.131204462364624e-27 | 2.7161211080176597e-28 | 5.432242216035319e-28 | 2.458028152052181e-30 | 1.2107545201743378e-30 |
| Bone marrow | 2.1149193020946 | 4.5092945713733025e-28 | 9.6484722629848e-27 | 5.579624813944677e-28 | 1.673887444183403e-27 | 7.57415133114662e-30 | 3.244096813937628e-30 |
| Go:0070469~respiratory chain | 7.252699903328269 | 1.9298186577523683e-26 | 2.7871279638493368e-24 | 9.897645304393964e-26 | 8.907880773954569e-25 | 1.9620882762014467e-27 | 1.0964878737229366e-28 |
| Respiratory chain | 7.638401642851031 | 2.285747408383908e-25 | 1.020097241281804e-23 | 6.12583003573042e-25 | 3.6754980214382524e-24 | 7.041183949115426e-27 | 3.734881386248216e-28 |
| Hippocampus | 2.2057288317386208 | 2.0519077193119936e-23 | 4.616992749592996e-22 | 2.002474066044195e-23 | 8.00989626417678e-23 | 3.6243874498537465e-25 | 1.476192603821578e-25 |
| Go:0031967~organelle envelope | 2.5774586076201875 | 7.082310457706249e-22 | 1.7380527361314114e-20 | 5.554953612865785e-22 | 5.554953612865785e-21 | 1.2235580645078822e-23 | 4.0240400327876416e-24 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Calcium | 24.42407660738714 | 0.28951832329015015 | 23.58258947236265 | 0.39447522789148326 | 0.39447522789148326 | 0.04094320600420348 | 0.001674146663500371 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0016126~sterol biosynthetic process | 26.64313725490196 | 2.9240576693290166e-07 | 1.555864814761776e-06 | 1.1067138742104632e-06 | 1.1067138742104632e-06 | 9.682541432131478e-10 | 2.6850860020339575e-11 |
| Go:0006695~cholesterol biosynthetic process | 30.89059391872691 | 1.4471084283096403e-06 | 6.469351243598709e-06 | 2.3008800749124703e-06 | 4.60175485583747e-06 | 4.026041486354809e-09 | 9.135795464339198e-11 |
| Steroid biosynthesis | 19.788916256157638 | 7.654530824741101e-07 | 1.7128864038262748e-05 | 3.3074079373029974e-06 | 3.3074079373029974e-06 | 1.3176945718181913e-08 | 5.360318265859517e-10 |
| Cholesterol biosynthesis | 34.20306513409962 | 6.466520103831641e-07 | 4.196162926017877e-05 | 4.051179080089007e-06 | 8.102341748084108e-06 | 3.22803762533391e-08 | 6.339727474138163e-10 |
| Go:0016125~sterol metabolic process | 11.53382565147271 | 4.62665943312679e-06 | 0.00029327155679492733 | 6.953411962529188e-05 | 0.000208587854230724 | 1.8251057976477485e-07 | 1.4020212486091872e-08 |
| Sterol biosynthesis | 25.65229885057471 | 1.061977756666721e-05 | 0.0002874902654048661 | 1.8503691473270578e-05 | 5.5510047266227325e-05 | 2.2116168074876236e-07 | 6.507251230132732e-09 |
| Go:0008203~cholesterol metabolic process | 11.418487394957983 | 0.00030200919567169304 | 0.0017197369964194742 | 0.00030577579485302486 | 0.0012225423007418135 | 1.0702445850094158e-06 | 8.3210687855148e-08 |
| Go:0006694~steroid biosynthetic process | 11.257663628831814 | 0.00021799907330632973 | 0.0019199139813519395 | 0.00027309915715822974 | 0.0013647501579526766 | 1.1948219262745415e-06 | 9.438217675387581e-08 |
| Mmu00900:terpenoid backbone biosynthesis | 30.136554621848738 | 8.242006913039557e-05 | 0.01671007077328479 | 0.0013069362450626176 | 0.0013069362450626176 | 1.556881965092711e-05 | 3.712768043136717e-07 |
| Lipid synthesis | 7.995521719659651 | 0.0003882128355635617 | 0.02155967411564541 | 0.0010403103177206718 | 0.004154752299853515 | 1.6587142369384825e-05 | 1.9033717653895786e-06 |
| Go:0008202~steroid metabolic process | 5.516177485486948 | 0.022582019768334516 | 0.13068541509396736 | 0.015383720974197157 | 0.08882442170579996 | 8.137872610876524e-05 | 1.3842872874142951e-05 |
| Go:0005576~extracellular region | 1.8478817733990147 | 0.026162617706394165 | 0.3579307591001446 | 0.054704796260282684 | 0.054704796260282684 | 0.0002869895012478547 | 0.0001506190416626758 |
| Secreted | 1.9200513425379866 | 0.06524543023697305 | 0.85151591623569 | 0.03248529455684113 | 0.1522108125880075 | 0.0006576453052594159 | 0.00033068686193789336 |
| Mmu00100:steroid biosynthesis | 19.854671280276815 | 0.008452431025824492 | 0.9836432780837567 | 0.037940599284707055 | 0.07444170949533135 | 0.0009205066240207212 | 3.8235104138517954e-05 |
| Go:0043086~negative regulation of catalytic activity | 6.155180223904742 | 0.3441543224176695 | 1.498590804423383 | 0.14224317176515489 | 0.6583752807290402 | 0.0009392283393975215 | 0.0001420201381923461 |
| Amidation | 10.993842364532021 | 0.1157210390722131 | 1.3650779717944483 | 0.04326907691474291 | 0.23309977052302566 | 0.001056805987548558 | 8.61186301386443e-05 |
| Mammary gland | 1.7047098579650457 | 0.0821862497730147 | 1.2053371838274929 | 0.10587540550649166 | 0.10587540550649166 | 0.0010859162383629133 | 0.0006167938440563743 |
| Lipoprotein | 2.538476335443726 | 0.08208289300251947 | 1.4397434016579003 | 0.039213413067430314 | 0.24423196385307266 | 0.0011149985656060193 | 0.00041975594559205595 |
| Go:0008610~lipid biosynthetic process | 3.4277720444903106 | 0.22257638515326472 | 2.1925393203871213 | 0.17890624932331045 | 0.7933944546584548 | 0.0013787021555322348 | 0.0003813967714577683 |
| Go:0008299~isoprenoid biosynthetic process | 16.147355912061794 | 0.8209842924987515 | 2.8660796281194223 | 0.2053321557091322 | 0.8736222091500128 | 0.0018080568588869398 | 9.653191319909874e-05 |
| Go:0044092~negative regulation of molecular function | 4.7096454743513565 | 0.5139011802853707 | 5.7208937533889115 | 0.34232313267924885 | 0.9848600073095138 | 0.0036594431628636705 | 0.0007283642532830037 |
| Go:0001558~regulation of cell growth | 5.791986359761296 | 0.7465005958921818 | 5.7736625209307 | 0.3192548313097099 | 0.9854510970876952 | 0.0036941568550092852 | 0.0005939334246671077 |
| Go:0042401~biogenic amine biosynthetic process | 12.249718278115845 | 0.9121769625343038 | 6.31545721076373 | 0.3207064365253516 | 0.9903463845594489 | 0.004051632346761579 | 0.0002947967118827307 |
| Cleavage on pair of basic residues | 3.325868278345821 | 0.2857428895740858 | 7.304285133653399 | 0.1672889096979705 | 0.7688175882345608 | 0.005817863763917164 | 0.0016482102210353706 |
| Go:0042423~catecholamine biosynthetic process | 22.202614379084967 | 0.999700488962424 | 11.570272281947247 | 0.4897263672905846 | 0.9998409840603655 | 0.007623032496677321 | 0.0002858421304630846 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0008203~cholesterol metabolic process | 43.13650793650793 | 0.006770042181034941 | 0.12047551633553866 | 0.03863378070701218 | 0.03863378070701218 | 8.454548405785008e-05 | 1.8713654324794746e-06 |
| Go:0016125~sterol metabolic process | 39.21500721500721 | 0.0009057965338234997 | 0.16009454373048237 | 0.02584373517336036 | 0.051019571699009836 | 0.00011236944591402652 | 2.746078094347024e-06 |
| Go:0006695~cholesterol biosynthetic process | 98.46376811594202 | 0.052433203795093086 | 0.5220268442514775 | 0.05542662842260915 | 0.15723382861546886 | 0.00036702648385376056 | 3.400110835582468e-06 |
| Go:0016126~sterol biosynthetic process | 75.48888888888888 | 0.08093088628256806 | 0.8914046831398292 | 0.07055075425884205 | 0.2537184286928722 | 0.0006278120893068637 | 7.749644360961088e-06 |
| Go:0008202~steroid metabolic process | 18.75500345065562 | 0.08077344029092604 | 1.3919475029796735 | 0.08755530185284932 | 0.36754044219969506 | 0.0009826477878556355 | 5.1042732724211055e-05 |
| Go:0005576~extracellular region | 3.525563909774436 | 0.06172060326530582 | 1.2549138263028836 | 0.0644083650559365 | 0.0644083650559365 | 0.0013045637112290552 | 0.00036190898380903037 |
| Secreted | 4.02343661971831 | 0.09645815070235075 | 2.1506083422105693 | 0.14812415513548371 | 0.14812415513548371 | 0.0020532031958734685 | 0.000497096264205239 |
| Go:0006694~steroid biosynthetic process | 31.896713615023472 | 0.2162049393995359 | 4.848861087065226 | 0.2371940059853722 | 0.8029916734238893 | 0.00348000213315566 | 0.00010545231617959868 |
| Signal | 2.404579124579125 | 0.6133325484997391 | 11.57647803306725 | 0.3646729271278495 | 0.5963595104757051 | 0.011563782015314664 | 0.004646964655259916 |
| Go:0030182~neuron differentiation | 7.567808409913672 | 0.41355908250151585 | 16.514620242234113 | 0.569484523740097 | 0.9972589221055929 | 0.012579868854123065 | 0.00161591528371096 |
| Calcium | 4.884815321477428 | 0.463450017887996 | 15.151979844311224 | 0.3322667888961537 | 0.7022793682068454 | 0.01541330968311292 | 0.0030472865778723096 |
| Protein kinase inhibitor | 119.02666666666666 | 0.29658311050361585 | 15.700673304326319 | 0.2701089754525148 | 0.7161871252470218 | 0.01601700992249913 | 0.00012317362779299141 |
| Go:0045596~negative regulation of cell differentiation | 12.443223443223443 | 0.8075389878235838 | 26.394469607873962 | 0.7140672238518915 | 0.9999553201684445 | 0.02126419721658905 | 0.001663122224322623 |
| Cholesterol biosynthesis | 79.35111111111112 | 0.25158249921447984 | 22.60419237131366 | 0.31469797309649716 | 0.8488493282542923 | 0.023933030252658385 | 0.00028407170339127003 |
| Mmu00900:terpenoid backbone biosynthesis | 74.51948051948051 | 0.06449931898397887 | 17.991733492528738 | 0.45729752203337015 | 0.45729752203337015 | 0.024151335325017297 | 0.0003002855597598153 |
| Go:0008299~isoprenoid biosynthetic process | 68.62626262626262 | 0.998790756265691 | 32.49609436751031 | 0.7600062581484128 | 0.999997358812376 | 0.027186447723290458 | 0.00037690763527829633 |
| Medulla oblongata | 5.800029192818567 | 0.47332035116013893 | 22.276098068108396 | 0.6459918247162426 | 0.6459918247162426 | 0.027675631910248962 | 0.004602059879481688 |
| Sterol biosynthesis | 59.51333333333333 | 0.5649193323922455 | 28.944406009150867 | 0.3429234145737635 | 0.9195185208308523 | 0.03178800018257759 | 0.0005098110697433965 |
| Go:0055092~sterol homeostasis | 58.06837606837606 | 0.9973486297391339 | 37.15538931254573 | 0.7808906789752035 | 0.9999997449598643 | 0.03205408512495799 | 0.0005286194732727484 |
| Go:0042632~cholesterol homeostasis | 58.06837606837606 | 0.9973486297391339 | 37.15538931254573 | 0.7808906789752035 | 0.9999997449598643 | 0.03205408512495799 | 0.0005286194732727484 |
| Go:0031225~anchored to membrane | 9.537757437070937 | 0.4664423016138125 | 29.19882017212283 | 0.5975465689836086 | 0.8380312358631347 | 0.03506368253455502 | 0.0035628881579055235 |
| Go:0043068~positive regulation of programmed cell death | 9.058666666666666 | 0.9332485840093009 | 42.64250518600243 | 0.8082623943413858 | 0.9999999871240198 | 0.03823670601012191 | 0.004092783107823523 |
| Go:0010942~positive regulation of cell death | 8.986772486772487 | 0.7494340713031286 | 43.11589587566408 | 0.7848811077051574 | 0.9999999901792956 | 0.03879558546124365 | 0.004185257310971446 |
| Go:0030888~regulation of b cell proliferation | 47.18055555555555 | 0.9986760438717217 | 43.55057044060887 | 0.7625154135511734 | 0.999999992357058 | 0.039312578977366994 | 0.0008029660510248117 |
| Cholesterol metabolism | 47.61066666666667 | 0.38701632227623817 | 34.767111278026675 | 0.36238920370107797 | 0.9571560370798997 | 0.03958237023108938 | 0.0007993812562580089 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.3581591870515088 | 4.7781785694756e-07 | 4.415172816241153e-06 | 1.1216283088977264e-06 | 1.1216283088977264e-06 | 3.232360067305254e-09 | 2.342244982975044e-09 |
| Brain | 1.3296120325221246 | 9.964594506195468e-07 | 1.181010054196463e-05 | 1.6537341350764123e-06 | 1.6537341350764123e-06 | 9.670967822341175e-09 | 7.168776600975159e-09 |
| Membrane | 1.3691549400726493 | 6.311701156613836e-06 | 5.888326443104219e-05 | 7.4793158745256605e-06 | 1.495857580879889e-05 | 4.310860917122128e-08 | 3.093980870315021e-08 |
| Glycoprotein | 1.432508853474989 | 0.000287177299924668 | 0.0028130082801447465 | 0.00023818018444254907 | 0.0007143703774387111 | 2.0594378261937113e-06 | 1.4079330442078817e-06 |
| Head | 1.7096058875582423 | 0.0004962658191327574 | 0.007709178883608114 | 0.000539622554804331 | 0.001078953917106995 | 6.3130584755321755e-06 | 3.5711375066840468e-06 |
| Go:0042995~cell projection | 2.2431493708733026 | 0.0008228073499486621 | 0.014876002213748407 | 0.0034749670192630866 | 0.0034749670192630866 | 1.1050792085321833e-05 | 4.676955209095389e-06 |
| Mmu05221:acute myeloid leukemia | 6.06425702811245 | 0.0009275941571477153 | 0.03527624374285443 | 0.004300138641810891 | 0.004300138641810891 | 2.9719633043268618e-05 | 4.180282428502118e-06 |
| Mmu05200:pathways in cancer | 2.568391211906449 | 0.001715198154799702 | 0.0502972864833362 | 0.0030677202512674473 | 0.006126029594994753 | 4.23775206761046e-05 | 1.5465383034119154e-05 |
| Go:0005886~plasma membrane | 1.4055062443674484 | 0.005169993413639551 | 0.05708574197867966 | 0.0066582426788615034 | 0.013272153162152356 | 4.2415022121777113e-05 | 2.9450725816425853e-05 |
| Alternative splicing | 1.3096064747926393 | 0.00919325664453663 | 0.0826286090906958 | 0.005236148117343409 | 0.020780662478094847 | 6.0515803233370305e-05 | 4.52723840948206e-05 |
| Go:0044459~plasma membrane part | 1.5577426186620158 | 0.013426821982680859 | 0.16643696356660742 | 0.012908027447046888 | 0.038226381522456276 | 0.00012372624952171416 | 7.680260070465369e-05 |
| Go:0016337~cell-cell adhesion | 2.6918776139115126 | 0.12636463293275435 | 0.6785710513359744 | 0.5521164472120776 | 0.5521164472120776 | 0.0003952083054654495 | 0.0001364474483118469 |
| Go:0035295~tube development | 2.540062967335695 | 0.118344345517715 | 0.889918924346611 | 0.40977849984139725 | 0.6516385807505285 | 0.0005188194891772874 | 0.0001908207514435265 |
| Lung | 1.5712478072067406 | 0.07923425999819522 | 1.1667612049536258 | 0.05330625859713323 | 0.15154557695774773 | 0.0009605846031305227 | 0.0005937059101766707 |
| Metal-binding | 1.3526119236699734 | 0.13665108307757956 | 1.3593020209399342 | 0.06717438754751948 | 0.2936786893572866 | 0.001001472343666137 | 0.0007200169507066707 |
| Embryonic tail | 2.1686456524008846 | 0.061928578080082786 | 1.27078778144335 | 0.04378385337238555 | 0.16396532271373387 | 0.001046733585675848 | 0.0004598164829391449 |
| Mmu05220:chronic myeloid leukemia | 4.093373493975903 | 0.09760556234292328 | 1.6604838473034178 | 0.06589979623458297 | 0.18495722779673496 | 0.001409451897936258 | 0.00030837154942603445 |
| Go:0048732~gland development | 2.687322829454809 | 0.2784617407374804 | 2.439570383626033 | 0.6213699993841639 | 0.945719346640493 | 0.00143282459575849 | 0.0004943775653972798 |
| Vagina | 2.720499202879415 | 0.09403338742367051 | 2.4789802087961554 | 0.0678858019843318 | 0.2963679450806064 | 0.0020534426481275897 | 0.0007101996677350427 |
| Go:0048589~developmental growth | 3.5293506493506497 | 0.7071707960256741 | 3.5026911514819403 | 0.6505990824328843 | 0.985096229146801 | 0.0020678104179423593 | 0.000531532176530624 |
| Go:0010604~positive regulation of macromolecule metabolic process | 1.7841899017254468 | 0.31095752874064375 | 3.5186899283807582 | 0.5705028684051485 | 0.9853849152306832 | 0.0020774164805314084 | 0.0011280068607714586 |
| Go:0043086~negative regulation of catalytic activity | 3.4944065835154943 | 0.8190222024873085 | 3.747450031876376 | 0.5280849591249017 | 0.9889545626574422 | 0.002214934077854517 | 0.0005753835349913063 |
| Zinc | 1.3954743063799944 | 0.3284940510023183 | 3.7500052825101826 | 0.14941303265843475 | 0.6212851410161777 | 0.002794278077471852 | 0.0019502154801517278 |
| Mmu05222:small cell lung cancer | 3.65995747696669 | 0.14652664843560137 | 3.3790377595907084 | 0.09964188387438655 | 0.3428551099311612 | 0.002891334445505368 | 0.0007134433951410144 |
| Brain cortex | 1.8199311412236647 | 0.18967409630396137 | 3.483687381339051 | 0.0794203570415869 | 0.3913491933682508 | 0.0028993592647546754 | 0.001511940483266496 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Lung | 2.7312647289866456 | 0.06704424306126211 | 1.5031652255173222 | 0.11177166186321408 | 0.11177166186321408 | 0.0014100339131791501 | 0.0004991379753239735 |
| Retina | 3.716887004240459 | 0.17212638826522386 | 2.779261684986234 | 0.10442408415348592 | 0.1979437789556776 | 0.0026224668876202394 | 0.0006792468299472896 |
| Go:0046903~secretion | 6.047622579927306 | 0.13958283957155837 | 4.276267189942096 | 0.8807113378161896 | 0.8807113378161896 | 0.00287300841380306 | 0.00045546575026453144 |
| Go:0010604~positive regulation of macromolecule metabolic process | 3.167119881905058 | 0.4719750148172219 | 9.277807027119556 | 0.9063785034944212 | 0.9912350153920559 | 0.006389500470243979 | 0.0019333156300048407 |
| Go:0032940~secretion by cell | 5.98801339679182 | 0.8540935030404398 | 13.006512659601277 | 0.8956071715369057 | 0.9988623412953797 | 0.009130970431326383 | 0.0014571110890197261 |
| Ubl conjugation pathway | 3.397618529795563 | 0.8410244416554853 | 17.657280086605553 | 0.918527826978874 | 0.918527826978874 | 0.016047228030312777 | 0.004497221806217048 |
| Go:0044265~cellular macromolecule catabolic process | 2.92616221163423 | 0.844706295930214 | 23.290173177172868 | 0.9602358433875094 | 0.9999974998440309 | 0.01730342755560722 | 0.005627852726285052 |
| Brain | 1.3584028442499658 | 0.828199094528407 | 17.35847087015785 | 0.3918564965545951 | 0.7750851062620507 | 0.01760548019654899 | 0.012592126413109205 |
| Go:0012501~programmed cell death | 3.2965722801788377 | 0.8147782715358787 | 23.70718998009528 | 0.9281281929934497 | 0.9999980822463398 | 0.017656016168429812 | 0.005096669616963853 |
| Mammary tumor metastatized to lung. tumor arose spontaneously | 3.872904483430799 | 0.46954331754185585 | 18.16347667943764 | 0.32440342309462167 | 0.791670984397242 | 0.018500962347696152 | 0.004550886417828652 |
| Go:0010817~regulation of hormone levels | 6.961065573770492 | 0.8194112831539897 | 25.18331938064613 | 0.9048670226769642 | 0.9999992587127456 | 0.01891872108844943 | 0.0025898724723145495 |
| Go:0005783~endoplasmic reticulum | 2.5825224894437304 | 0.71426405603637 | 20.06303589933833 | 0.9082543953812396 | 0.9082543953812396 | 0.01923325016580156 | 0.00709227101968542 |
| Membrane | 1.407734653503197 | 0.9407049387888873 | 21.55253408293619 | 0.7912210279526645 | 0.9564113408308579 | 0.02000972812116675 | 0.013753704568267081 |
| Amino acid transport and metabolism | 11.333333333333334 | 0.057399694221968 | 9.21396214360033 | 0.10518005650783491 | 0.10518005650783491 | 0.021981362402258935 | 0.001845574207526105 |
| Go:0034754~cellular hormone metabolic process | 12.851197982345523 | 0.9998153586746747 | 28.753972449893595 | 0.9052279425970905 | 0.9999999313307725 | 0.022072069793283394 | 0.0016269129045930087 |
| Go:0008219~cell death | 3.075500371843373 | 0.9133057312112176 | 30.779379652246885 | 0.893235681692951 | 0.9999999831185771 | 0.02392703135886169 | 0.007382816675381431 |
| Go:0043632~modification-dependent macromolecule catabolic process | 3.0694462372531306 | 0.9928599436716309 | 30.99934437383528 | 0.8654428431925664 | 0.9999999855402356 | 0.02413152807548348 | 0.007459966518373997 |
| Go:0019941~modification-dependent protein catabolic process | 3.0694462372531306 | 0.9928599436716309 | 30.99934437383528 | 0.8654428431925664 | 0.9999999855402356 | 0.02413152807548348 | 0.007459966518373997 |
| Go:0009057~macromolecule catabolic process | 2.724820775053893 | 0.9408471589655563 | 31.400655476693895 | 0.8401576783658788 | 0.9999999891127602 | 0.024506192904965394 | 0.00853197211862593 |
| Go:0016265~death | 3.0043905366562433 | 0.9371292938847037 | 33.457769472286046 | 0.8349547330415545 | 0.999999997524666 | 0.02645946762337192 | 0.008348816897528397 |
| Mammary gland | 1.8461938745168005 | 0.8518783653402833 | 25.012505139331797 | 0.362706074639773 | 0.8948767221471102 | 0.026460542230752622 | 0.01364505414360692 |
| Go:0010557~positive regulation of macromolecule biosynthetic process | 2.942035261367151 | 0.9544075032507524 | 35.98081366275753 | 0.8360375954872212 | 0.9999999996224875 | 0.028933678458862737 | 0.009313966279593723 |
| Go:0003006~reproductive developmental process | 4.218827620466965 | 0.9868360908784486 | 36.1168724031748 | 0.8130649550344218 | 0.999999999659608 | 0.02906967809204842 | 0.006539548246939959 |
| Go:0051603~proteolysis involved in cellular protein catabolic process | 2.91999754405354 | 0.959703283642204 | 36.91238311757711 | 0.7982555546455103 | 0.9999999998149784 | 0.029870301059457233 | 0.009684573146072793 |
| Go:0044257~cellular protein catabolic process | 2.9036847086118995 | 0.9633547208629271 | 37.61550746747797 | 0.78354819858283 | 0.9999999998927449 | 0.030585834813470575 | 0.009969579042560638 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Hippocampus | 8.048069133135295 | 0.46450163289220203 | 23.78821387007205 | 0.47155495242625456 | 0.47155495242625456 | 0.03682351209982295 | 0.004483139574752129 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Ribosomal protein | 17.120147203648994 | 1.205349850272513e-32 | 8.127207949903237e-31 | 1.5429039646465847e-31 | 1.5429039646465847e-31 | 6.271967335961727e-34 | 2.9542888487071395e-35 |
| Go:0005840~ribosome | 14.428892215568863 | 1.1627598688427322e-30 | 1.4545498578041348e-28 | 2.3500593199168885e-29 | 2.3500593199168885e-29 | 1.1576646896142309e-31 | 6.60659016387916e-33 |
| Ribonucleoprotein | 12.315651514106367 | 7.764667532367217e-30 | 7.031251575219252e-28 | 6.674214563417497e-29 | 1.3348429126834994e-28 | 5.426190701965445e-31 | 3.8062095746898126e-32 |
| Mmu03010:ribosome | 17.526344496563762 | 1.2756825464618105e-26 | 3.062469119623133e-24 | 2.44240785318366e-25 | 2.44240785318366e-25 | 2.840009131608907e-27 | 1.1492635553710004e-28 |
| Acetylation | 3.240737890894742 | 9.923341323628032e-25 | 2.1000915442528695e-23 | 1.3289679103088531e-24 | 3.9869037309265594e-24 | 1.6206925735473818e-26 | 4.864383001778447e-27 |
| Go:0006412~translation | 9.551743136696114 | 2.790838346112325e-23 | 7.074184283685347e-22 | 4.340415914267004e-22 | 4.340415914267004e-22 | 4.4978403256652885e-25 | 4.2285429486550373e-26 |
| Mitochondrion | 5.183486238532111 | 4.2077764148823037e-20 | 1.4590833091890425e-18 | 6.924965610496762e-20 | 2.7699862441987046e-19 | 1.1260106683734571e-21 | 2.0626354974913253e-22 |
| Go:0030529~ribonucleoprotein complex | 6.482619177229956 | 1.2537903254472085e-19 | 6.2532528627617184e-18 | 5.051568047352899e-19 | 1.0103136094705798e-18 | 4.976914332367388e-21 | 7.123808667313685e-22 |
| Kidney | 3.1526884345437898 | 2.7070356044256002e-19 | 6.998064541582636e-18 | 6.356075360275372e-19 | 6.356075360275372e-19 | 6.293143921064724e-21 | 1.9475076290831656e-21 |
| Go:0005739~mitochondrion | 3.454863344415556 | 4.4162424700819886e-17 | 1.1258611735360257e-15 | 6.06336622427689e-17 | 1.8190098672830666e-16 | 8.960639740310673e-19 | 2.509228676182948e-19 |
| Mitochondrion inner membrane | 11.18839925803379 | 5.469856922238116e-17 | 4.419717275507037e-15 | 1.6781161129457746e-16 | 8.3905805647288725e-16 | 3.4108051076133627e-18 | 2.6813024128618217e-19 |
| Go:0031966~mitochondrial membrane | 6.714267117938037 | 2.739018960288069e-16 | 1.4188325829418998e-14 | 5.7308807894321e-16 | 2.29235231577284e-15 | 1.1292375939767685e-17 | 1.5562607728909482e-18 |
| Mmu00190:oxidative phosphorylation | 10.28469006721434 | 3.2578091608002535e-16 | 1.942163872338482e-14 | 7.744659796851825e-16 | 1.548931959370365e-15 | 1.801083673686471e-17 | 1.4674816039640781e-18 |
| Go:0005743~mitochondrial inner membrane | 7.588606570642498 | 3.90062641017342e-16 | 2.3115633631088154e-14 | 7.469405047762614e-16 | 3.734702523881307e-15 | 1.839754937872565e-17 | 2.216265005780352e-18 |
| Go:0005740~mitochondrial envelope | 6.319310228647565 | 1.7152889512461378e-15 | 1.4432899320127035e-13 | 3.774758283725532e-15 | 2.2537527399890678e-14 | 6.618695607606348e-17 | 9.745959950262147e-18 |
| Go:0019866~organelle inner membrane | 7.199447259327499 | 1.7122546224057488e-15 | 1.4432899320127035e-13 | 3.219646771412954e-15 | 2.2537527399890678e-14 | 7.619365260127722e-17 | 9.72871944548721e-18 |
| Go:0044429~mitochondrial part | 5.286922338529049 | 4.942261850637198e-15 | 1.4432899320127035e-13 | 2.7755575615628914e-15 | 2.2537527399890678e-14 | 1.5729802887774084e-16 | 2.8081033242256806e-17 |
| Respiratory chain | 21.135247114530927 | 2.2648549702353193e-14 | 8.659739592076221e-13 | 2.731148640577885e-14 | 1.638689184346731e-13 | 6.228839391203385e-16 | 2.2616051625047645e-17 |
| Go:0070469~respiratory chain | 18.430584983878397 | 3.907985046680551e-14 | 5.295763827461997e-12 | 9.514611321037592e-14 | 8.564260411958458e-13 | 4.265841698572746e-15 | 1.757413172574921e-16 |
| Go:0033279~ribosomal subunit | 18.151333696243878 | 1.1723955140041653e-13 | 6.972200594645983e-12 | 1.1268763699945339e-13 | 1.1268763699945339e-12 | 5.533599138166325e-15 | 2.29162495740566e-16 |
| Mmu05012:parkinson's disease | 8.796116504854368 | 8.626432901337466e-13 | 8.619771563189715e-11 | 2.291500322826323e-12 | 6.874500968478969e-12 | 7.992501409772402e-14 | 7.792534964640273e-15 |
| Go:0031967~organelle envelope | 4.714304723885562 | 4.083844373781176e-12 | 1.453503983839255e-10 | 2.134958876354176e-12 | 2.3484103550686086e-11 | 1.1565464003806332e-13 | 2.3241487636585868e-14 |
| Go:0031975~envelope | 4.6969087654948405 | 4.552802579382842e-12 | 1.61259894326804e-10 | 2.171152146956956e-12 | 2.6053381674273624e-11 | 1.283812947205531e-13 | 2.589127519475532e-14 |
| Mmu05016:huntington's disease | 6.6972253169929425 | 6.160505439112285e-11 | 4.473765802259777e-09 | 8.91990925566688e-11 | 3.567961481820703e-10 | 4.1488346181971706e-12 | 5.550123361710903e-13 |
| Go:0006091~generation of precursor metabolites and energy | 6.941507024265644 | 7.650979849671558e-10 | 9.121559063629547e-09 | 2.7982988148878007e-09 | 5.5965976297756015e-09 | 5.799626552106969e-12 | 7.727876867485767e-13 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.6306044745644197 | 3.003243602924133e-58 | 3.535018241581146e-57 | 1.0591051196170607e-57 | 1.0591051196170607e-57 | 2.515689120230548e-60 | 1.4721782367275162e-60 |
| Alternative splicing | 1.6648187567251103 | 9.610454456610287e-38 | 1.1578869261227525e-36 | 1.73453697772965e-37 | 3.4690739554593e-37 | 8.240080654297624e-40 | 4.7110070865736705e-40 |
| Brain | 1.3975875416802532 | 6.769791076890664e-26 | 8.93311854072387e-25 | 1.5732958211735229e-25 | 1.5732958211735229e-25 | 6.992425871882324e-28 | 4.870353292727096e-28 |
| Nucleus | 1.4789647072701912 | 4.738047849909527e-16 | 4.99996293693737e-15 | 4.993360696244813e-16 | 1.4980082088734437e-15 | 3.558214272858536e-18 | 2.3225724754458467e-18 |
| Transcription | 1.666838603548871 | 1.857181075592962e-11 | 2.1934676297519218e-10 | 1.642930236300799e-11 | 6.571698740742704e-11 | 1.5612713282903114e-13 | 9.10192910289378e-14 |
| Zinc-finger | 1.8245306725997297 | 4.3236081381792246e-11 | 5.945466341472638e-10 | 3.56255025479868e-11 | 1.7812762376223645e-10 | 4.2309629701580737e-13 | 2.119877921620209e-13 |
| Transcription regulation | 1.7070055325108455 | 5.659872570618063e-11 | 7.171707672171124e-10 | 3.58109097930992e-11 | 2.1486556978089766e-10 | 5.103434668865436e-13 | 2.774923274833442e-13 |
| Embryonic tail | 2.6271709612226855 | 4.097222561227909e-11 | 1.1249001730107011e-09 | 9.905831710455004e-11 | 1.9811652318679762e-10 | 8.805316566605982e-13 | 2.9478121685005e-13 |
| Coiled coil | 1.6513731899466593 | 1.1129497323736359e-10 | 1.3488432593078414e-09 | 5.773104216899583e-11 | 4.0411740620527326e-10 | 9.599313456007085e-13 | 5.455942484662668e-13 |
| Go:0006350~transcription | 1.6208474215018553 | 2.3385626768401835e-10 | 2.230016171722582e-09 | 3.439026841078885e-09 | 3.439026841078885e-09 | 1.2505641323652245e-12 | 7.086868456027394e-13 |
| Zinc | 1.602520374824974 | 4.027365108072445e-10 | 4.6394998953758204e-09 | 1.7375156868837394e-10 | 1.390012438484689e-09 | 3.301699109627548e-12 | 1.974168360508631e-12 |
| Thymus | 1.525601186437193 | 6.305093114278293e-10 | 9.158329650205133e-09 | 5.376532552503477e-10 | 1.612959765751043e-09 | 7.1686794734128704e-12 | 4.536084358543286e-12 |
| Macrophage | 2.1931752067489505 | 7.791050027350366e-10 | 1.73419612004011e-08 | 7.635642118586361e-10 | 3.0542568474345444e-09 | 1.357448756255311e-11 | 5.605047794655796e-12 |
| Cerebellum | 1.6524423888347939 | 7.868951268363844e-09 | 1.2949710193055353e-07 | 4.561391264701342e-09 | 2.2806956101462106e-08 | 1.0136430042476676e-10 | 5.661112964660455e-11 |
| Brain cortex | 2.1400483396385037 | 6.680988207108385e-09 | 1.4233790812667735e-07 | 4.178078660821427e-09 | 2.5068471964928563e-08 | 1.1141538519530167e-10 | 4.8064650940714405e-11 |
| Metal-binding | 1.4402368051745402 | 2.4729067793494153e-08 | 2.5193978014925733e-07 | 8.386903282975311e-09 | 7.548212688224254e-08 | 1.7929242674467992e-10 | 1.2122092012157886e-10 |
| Go:0045449~regulation of transcription | 1.4664505647621564 | 9.98832200282962e-08 | 8.256457628696978e-07 | 6.366352918174556e-07 | 1.273270178292485e-06 | 4.630076077243541e-10 | 3.0267647848011877e-10 |
| Eye | 1.611065587763324 | 2.544124735326392e-07 | 3.869000209100193e-06 | 9.734373207503921e-08 | 6.814059253512639e-07 | 3.028471768872539e-09 | 1.8303058186755242e-09 |
| Cytoplasm | 1.3579892037308494 | 2.3950651510951104e-06 | 2.3491671352626753e-05 | 7.038193826636174e-07 | 7.038171535245219e-06 | 1.6717805809954055e-08 | 1.1740529385189164e-08 |
| Go:0051276~chromosome organization | 2.1652572982620146 | 6.294359964575946e-06 | 4.0727845340704505e-05 | 2.0935956603573835e-05 | 6.280655487711506e-05 | 2.2839464174916128e-08 | 9.536938971209524e-09 |
| Go:0031981~nuclear lumen | 1.7701019252548131 | 3.6566322570985577e-06 | 5.462277701262508e-05 | 1.6265243472979307e-05 | 1.6265243472979307e-05 | 3.8912381379378537e-08 | 2.0776357406686026e-08 |
| Go:0043228~non-membrane-bounded organelle | 1.4660760812923397 | 1.755995567753743e-05 | 0.00010745080292196008 | 1.5998034716857568e-05 | 3.199581349644376e-05 | 7.654623005931138e-08 | 4.988667450891444e-08 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.4660760812923397 | 1.755995567753743e-05 | 0.00010745080292196008 | 1.5998034716857568e-05 | 3.199581349644376e-05 | 7.654623005931138e-08 | 4.988667450891444e-08 |
| Head | 1.5180082942605573 | 7.234211209294905e-06 | 0.00010378246212416897 | 2.2847621945487617e-06 | 1.827795139297539e-05 | 8.123607865637387e-08 | 5.204487191196262e-08 |
| Dna-binding | 1.543624326861321 | 1.0164224071740335e-05 | 0.00011676199467292037 | 3.180209946274104e-06 | 3.4981753158302986e-05 | 8.309350017145678e-08 | 4.982487981812784e-08 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Ribosomal protein | 10.61003557229546 | 9.568304424133392e-52 | 5.50401036012198e-50 | 1.5446617272078983e-50 | 1.5446617272078983e-50 | 3.9606710954048673e-53 | 2.3451726529738706e-54 |
| Ribonucleoprotein | 7.954998607630187 | 1.1950717056556572e-47 | 8.864250645715254e-46 | 1.2438447438270087e-46 | 2.4876894876540174e-46 | 6.37869099398466e-49 | 5.858194635566947e-50 |
| Go:0005840~ribosome | 8.85799808429119 | 9.769027125179685e-47 | 1.009367436269236e-44 | 2.513806936220816e-45 | 2.513806936220816e-45 | 7.415359693866715e-48 | 5.550583593852093e-49 |
| Go:0005739~mitochondrion | 2.9172448571478253 | 1.2218517243239661e-36 | 3.0056451788218557e-35 | 3.7427459153366584e-36 | 7.485491830673317e-36 | 2.2081096845644004e-38 | 6.942339342749808e-39 |
| Mitochondrion | 3.9173333333333336 | 2.3794593197439852e-36 | 7.161498948914124e-35 | 6.699414070859466e-36 | 2.0098242212578396e-35 | 5.153395439122666e-38 | 1.166401627325483e-38 |
| Mmu03010:ribosome | 10.377327997537325 | 3.088021870228702e-36 | 6.725840276258318e-34 | 8.168151926348532e-35 | 8.168151926348532e-35 | 5.672327726630925e-37 | 2.7820016848907223e-38 |
| Kidney | 2.5266226461237467 | 3.867918274576191e-34 | 9.110537847577065e-33 | 1.2881183681396731e-33 | 1.2881183681396731e-33 | 7.445770914102156e-36 | 2.782675017680713e-36 |
| Go:0006412~translation | 6.025312401845157 | 1.150348819778086e-33 | 2.211791167008176e-32 | 2.373117608308521e-32 | 2.373117608308521e-32 | 1.3003384155115182e-35 | 1.7429527572395243e-36 |
| Mmu00190:oxidative phosphorylation | 8.011433087460485 | 1.9448992503280187e-33 | 1.3166949197561215e-31 | 7.995256877555849e-33 | 1.5990513755111698e-32 | 1.1104523441049792e-34 | 8.760807433909995e-36 |
| Mitochondrion inner membrane | 8.094102408419348 | 2.91512957425448e-33 | 2.211238583988308e-31 | 1.5514213214237253e-32 | 6.205685285694901e-32 | 1.591201355306385e-34 | 1.4289850854188628e-35 |
| Acetylation | 2.3435596973317403 | 6.633091813103906e-32 | 1.1174987631799404e-30 | 6.272363083444347e-32 | 3.1361815417221737e-31 | 8.041491132620958e-34 | 3.2515155946587773e-34 |
| Mmu05012:parkinson's disease | 7.682974559686888 | 5.824094718616864e-32 | 7.359608297830137e-30 | 2.979275759654445e-31 | 8.937827278963335e-31 | 6.206824499280093e-33 | 5.246932178934112e-34 |
| Go:0030529~ribonucleoprotein complex | 4.458973976215356 | 5.026060079307545e-31 | 2.035561521633159e-29 | 1.6898400880650618e-30 | 5.069520264195185e-30 | 1.495433706252267e-32 | 2.8557159541520142e-33 |
| Go:0005743~mitochondrial inner membrane | 5.5838769804287045 | 9.045883929409706e-31 | 4.860318286157659e-29 | 3.026128390158661e-30 | 1.2104513560634644e-29 | 3.570652967738833e-32 | 5.139706778073697e-33 |
| Mmu05016:huntington's disease | 6.22808593457594 | 4.4110066172085226e-30 | 4.046056448060356e-28 | 1.2284265761929686e-29 | 4.913706304771874e-29 | 3.4122960449804683e-31 | 3.9738798353229933e-32 |
| Go:0031966~mitochondrial membrane | 4.881934032983508 | 1.8480149747804702e-29 | 8.369873131804669e-28 | 4.1689962204024583e-29 | 2.084498110201229e-28 | 6.148961976994777e-31 | 1.0500085083979944e-31 |
| Go:0019866~organelle inner membrane | 5.2975243147656945 | 3.0172658843107825e-29 | 1.5148593185193216e-27 | 6.287871864857838e-29 | 3.7727231189147025e-28 | 1.1128976751960774e-30 | 1.7143556160856718e-31 |
| Go:0005740~mitochondrial envelope | 4.594761442808007 | 1.2259351095465294e-27 | 5.149545979429402e-26 | 1.832118376651542e-27 | 1.2824828636560792e-26 | 3.7831352910208826e-29 | 6.965540395150736e-30 |
| Go:0044429~mitochondrial part | 3.9313854523120124 | 7.410338962112572e-27 | 2.5773384981047117e-25 | 8.023504574151419e-27 | 6.418803659321135e-26 | 1.893452406879391e-28 | 4.2104198648366886e-29 |
| Respiratory chain | 13.651804062126642 | 1.3542803343035833e-26 | 8.07932883879231e-25 | 3.7790111882871205e-26 | 2.267406712972272e-25 | 5.81386336659557e-28 | 2.2128763632411494e-29 |
| Go:0070469~respiratory chain | 12.161273209549073 | 1.979447511886658e-26 | 3.514337335188755e-24 | 9.724864693553166e-26 | 8.75237822419785e-25 | 2.581822485014115e-27 | 1.1246860862992376e-28 |
| Go:0031967~organelle envelope | 3.5043848446147297 | 3.7164595113731064e-21 | 1.1282724340937743e-19 | 2.809937163472289e-21 | 2.809937163472289e-20 | 8.288900187233891e-23 | 2.111624722371083e-23 |
| Mmu05010:alzheimer's disease | 5.290569020021075 | 3.2933514452540745e-21 | 1.1998753850268656e-19 | 2.9143613392608512e-21 | 1.4571806696304254e-20 | 1.0119310205766844e-22 | 1.4834916420063398e-23 |
| Go:0031975~envelope | 3.4914535352250073 | 4.7242094006569024e-21 | 1.4280312334697073e-19 | 3.233163384264527e-21 | 3.5564797226909794e-20 | 1.049109062740702e-22 | 2.684209886736876e-23 |
| Go:0006091~generation of precursor metabolites and energy | 5.216358421967797 | 6.067747010007163e-20 | 6.475266976119891e-19 | 3.473784123189778e-19 | 6.947568246379556e-19 | 3.806886710344962e-22 | 6.12903738384562e-23 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.6810170417880903 | 4.537287189557526e-18 | 5.0347978911768724e-17 | 1.0951079616947765e-17 | 1.0951079616947765e-17 | 3.789300905518258e-20 | 2.224160387038003e-20 |
| Alternative splicing | 1.790083088337554 | 1.1324274851176597e-13 | 1.3322676295501878e-12 | 1.4432899320127035e-13 | 2.887690087050032e-13 | 1.0357552629467899e-15 | 5.47480009233993e-16 |
| Brain | 1.4396748092905622 | 4.8797225038477166e-08 | 6.050575707128303e-07 | 7.221519660038922e-08 | 7.221519660038922e-08 | 5.121645378612313e-10 | 3.510591982338984e-10 |
| Nucleus | 1.6579801485811716 | 2.073686501402605e-07 | 2.2854756531742737e-06 | 1.6570293770445943e-07 | 4.971087307348299e-07 | 1.7200998894711983e-09 | 1.0165130898428321e-09 |
| Macrophage | 3.0274418540220807 | 2.4138337734180837e-06 | 6.531424321387647e-05 | 3.897706270294954e-06 | 7.795397348409061e-06 | 5.528672003074626e-08 | 1.7365731418421066e-08 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.9363268998200711 | 2.4512807158649785e-05 | 0.00018004935612436768 | 3.4765165206396276e-05 | 3.4765165206396276e-05 | 1.3850903233269032e-07 | 6.963950780327491e-08 |
| Go:0043228~non-membrane-bounded organelle | 1.9363268998200711 | 2.4512807158649785e-05 | 0.00018004935612436768 | 3.4765165206396276e-05 | 3.4765165206396276e-05 | 1.3850903233269032e-07 | 6.963950780327491e-08 |
| Thymus | 1.7475970819996482 | 7.636469093352805e-05 | 0.0011620961169800381 | 4.623226685596116e-05 | 0.00013869038839919856 | 9.836875076436058e-07 | 5.494070926746917e-07 |
| Testis | 1.52855982588722 | 0.0007946045826547898 | 0.010556708626585642 | 0.0003149596907681129 | 0.0012592436903972049 | 8.936394140775985e-06 | 5.718835792759514e-06 |
| Go:0006974~response to dna damage stimulus | 3.4598231037255434 | 0.0008920455034728825 | 0.016570154120465475 | 0.015030304479792256 | 0.015030304479792256 | 9.956823706380355e-06 | 2.7043709275520076e-06 |
| Zinc | 1.7286827424039835 | 0.0020757617220473756 | 0.024119069735828447 | 0.0013108199403212284 | 0.005232979274096494 | 1.8154573145516306e-05 | 1.018582607217675e-05 |
| Go:0006281~dna repair | 3.7665973665973667 | 0.001902900626616577 | 0.03886056076711775 | 0.017603629132831977 | 0.03489737050701769 | 2.3353331180465042e-05 | 5.771842258357112e-06 |
| Dna damage | 4.0013446884805015 | 0.0011838459608043417 | 0.033005049807766085 | 0.001434977773842605 | 0.007154326784409748 | 2.4844126335610296e-05 | 5.80658735727654e-06 |
| Dna repair | 4.187919912875932 | 0.001430371963786481 | 0.04218286671251459 | 0.0015283374825111329 | 0.009135058979655719 | 3.175395773368585e-05 | 7.016622061408037e-06 |
| Zinc-finger | 1.93420482449805 | 0.0032334218756429056 | 0.04243950323860046 | 0.0013181138911183554 | 0.009190391377735807 | 3.194718369705825e-05 | 1.587566162768136e-05 |
| Coiled coil | 1.7329919335944037 | 0.004629316430834418 | 0.05404882285975354 | 0.0014688227596504344 | 0.011690350881643385 | 4.068851579194914e-05 | 2.274515750648729e-05 |
| Eye | 1.8466014924070848 | 0.003113634530785281 | 0.05063651766367139 | 0.0012082956287408075 | 0.006026895990638992 | 4.2872350687334226e-05 | 2.2434942406850124e-05 |
| Go:0045449~regulation of transcription | 1.5957721667645333 | 0.011835962737828498 | 0.09844092096849 | 0.029556924762488168 | 0.08607576016150453 | 5.9174843145066534e-05 | 3.607985031280787e-05 |
| Go:0005856~cytoskeleton | 1.9450105942891733 | 0.010650428840106363 | 0.15936567974702553 | 0.015280289458311724 | 0.030327091670693695 | 0.00012268758186859206 | 6.083650506770648e-05 |
| Cerebellum | 1.755440152908507 | 0.011116156798531285 | 0.17406379657847193 | 0.003459513666054348 | 0.020578384414071338 | 0.00014745770665161432 | 8.041693179295516e-05 |
| Cytoskeleton | 2.307922539587839 | 0.014332305575369353 | 0.22689301407812223 | 0.0054746346967388115 | 0.04820640423087008 | 0.00017094413778212005 | 7.076222709297218e-05 |
| Cytoplasm | 1.4522322859699237 | 0.03261608175005948 | 0.3213029030698644 | 0.006975399261722526 | 0.06760469839284522 | 0.0002421797038935286 | 0.000162535036874555 |
| Go:0033554~cellular response to stress | 2.587204874333587 | 0.06496046153816981 | 0.46162840329718735 | 0.10031677312319498 | 0.3448232228639301 | 0.00027796937447617 | 0.00010176219075961536 |
| Go:0000932~cytoplasmic mrna processing body | 14.745283018867926 | 0.01178368358973414 | 0.40068777964580304 | 0.025510381684774375 | 0.07459540796853226 | 0.0003088138223679247 | 1.6837445555280084e-05 |
| Liver | 1.4274049300136258 | 0.034207354215777475 | 0.43199378717999615 | 0.007354433322980358 | 0.050359012291006144 | 0.00036639579319581127 | 0.00025037239410328896 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Alternative splicing | 1.941107467998787 | 0.2195600170029296 | 2.570472423477288 | 0.18595321421856648 | 0.18595321421856648 | 0.0024175135458541944 | 0.0012144454809738512 |
| Coiled coil | 2.907473204239948 | 0.1671387590609682 | 2.8544719913421557 | 0.1080991416830166 | 0.2045128589334283 | 0.002688152147720108 | 0.000896109176673016 |
| Brain | 1.6022187393717546 | 0.42529817566762007 | 6.37534882975086 | 0.32939410794065827 | 0.32939410794065827 | 0.006528980824629918 | 0.003976991165519592 |
| Cytoskeleton | 4.711439503892334 | 0.26969500779951183 | 7.797621107517938 | 0.19249044665321968 | 0.47344588994547665 | 0.007517497431469672 | 0.0015394659349421496 |
| Go:0006281~dna repair | 9.067734401067733 | 0.258506766549056 | 10.168588422528213 | 0.8269136662582031 | 0.8269136662582031 | 0.008478247307390593 | 0.0009059204452697882 |
| Hippocampus | 3.370558867723329 | 0.4349268486314867 | 13.566292042427675 | 0.357348035084855 | 0.5869984519907033 | 0.01439221339713993 | 0.004098055389261411 |
| Go:0006974~response to dna damage stimulus | 7.014066331139502 | 0.5371044601302661 | 19.349618309197947 | 0.8277266567906376 | 0.9703218952194692 | 0.0169295507225393 | 0.0023313807254083966 |
| Cytoplasm | 1.96478485748872 | 0.8621721882550295 | 19.356312228776996 | 0.34617219071503647 | 0.8172516123292468 | 0.0197972329482028 | 0.00966742838612098 |
| Nucleus | 1.8032886231415644 | 0.8957422299826809 | 20.12948913797875 | 0.2989327950327836 | 0.8306449079588261 | 0.02067455550045303 | 0.011021600819586051 |
| Go:0006511~ubiquitin-dependent protein catabolic process | 10.707643814026794 | 0.9877769747721395 | 31.445377306972787 | 0.8723380501766878 | 0.9979194199981942 | 0.029531423170685918 | 0.002665794256520992 |
| Spinal ganglion | 5.158844530996429 | 0.6392896734078664 | 33.2507239118458 | 0.558368273346938 | 0.9138647742733085 | 0.03939698041428318 | 0.007308986275766059 |
| Go:0033554~cellular response to stress | 4.982764943160983 | 0.9945255331498289 | 40.89468661942649 | 0.8835419501775079 | 0.9998160592076301 | 0.040892318684465995 | 0.007859347075392133 |
| Go:0006259~dna metabolic process | 4.781560658045218 | 0.9975422780202291 | 44.23596533910447 | 0.8519993096536453 | 0.9999289901319267 | 0.04531352403672681 | 0.009062504471074316 |
| Embryonic lung | 40.7548717948718 | 0.1412216750393177 | 38.22227424547305 | 0.5182488026256966 | 0.9461369166040257 | 0.04676167411526009 | 0.0010946842232767917 |
| Mmu04010:mapk signaling pathway | 7.217610062893081 | 0.5230832408571295 | 35.78725006376134 | 0.8126423511459184 | 0.8126423511459184 | 0.04948332477874311 | 0.00664819253271615 |
| Dna repair | 7.938639395286795 | 0.7260030068132963 | 43.87751262053967 | 0.5326164061837133 | 0.9895758468859196 | 0.052273918241914026 | 0.006326170413135658 |
| Dna damage | 7.079302141157811 | 0.8304449328455583 | 50.90621833234752 | 0.5520015156627742 | 0.9963781053578117 | 0.06398760942918827 | 0.008661183394432458 |
| Cell projection | 7.0070643642072215 | 0.9740129381921874 | 51.556068271660635 | 0.511176804316879 | 0.9967400236663847 | 0.06514619638164454 | 0.008906561466550745 |
| Actin-binding | 6.0769230769230775 | 0.9318759656228287 | 60.788762092604706 | 0.5603779567393229 | 0.9993866404327432 | 0.08333971956732855 | 0.013082422343306995 |
| Phosphoprotein | 1.3782447415786032 | 0.9999959566290978 | 61.66629946414859 | 0.5311807727378326 | 0.9994870746693925 | 0.08526614138239802 | 0.05905883554742343 |
| Go:0042995~cell projection | 3.6243478260869564 | 0.9821104995602378 | 61.464489733457974 | 0.9989791150600512 | 0.9989791150600512 | 0.08663486109732794 | 0.022601696765671506 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain | 1.5093364936110734 | 1.0187346521917107e-09 | 1.3102330331804524e-08 | 1.4903289713430468e-09 | 1.4903289713430468e-09 | 1.1205475450175175e-11 | 7.329035914836671e-12 |
| Cerebellum | 1.9959815149688565 | 0.00027447015773796046 | 0.00476670885651842 | 0.00027106527169074024 | 0.0005420570669999458 | 4.076713743112281e-06 | 1.974874536020489e-06 |
| Mmu04012:erbb signaling pathway | 7.669072440523924 | 0.0001748232305394115 | 0.005074740772892383 | 0.0005012823525976096 | 0.0005012823525976096 | 4.47684744849266e-06 | 5.250404444201961e-07 |
| Brain cortex | 2.58495966692688 | 0.0021682830641264195 | 0.049290685767400255 | 0.0018675772368895727 | 0.005592274690280785 | 4.216429903633885e-05 | 1.5615972986043276e-05 |
| Mmu05211:renal cell carcinoma | 7.625249169435216 | 0.0018010426324012174 | 0.07794461777848039 | 0.003844639825187679 | 0.007674498394989904 | 6.878423158729033e-05 | 8.12008621977937e-06 |
| Go:0048666~neuron development | 3.3538195729976548 | 0.008083636568990915 | 0.1434861967214207 | 0.1122756329121275 | 0.1122756329121275 | 8.743669231203538e-05 | 2.459511059317189e-05 |
| Go:0030182~neuron differentiation | 2.9146289146289144 | 0.009964733877416143 | 0.15283511026296104 | 0.06145997196701791 | 0.1191426157798493 | 9.313776636055627e-05 | 3.034715812759551e-05 |
| Mmu00562:inositol phosphate metabolism | 8.649009474590871 | 0.007352202308436584 | 0.14651364559191693 | 0.00481718614397697 | 0.014382054369058261 | 0.00012933519392044275 | 1.329606107826193e-05 |
| Go:0045202~synapse | 3.1674741141825784 | 0.00839187818152698 | 0.20693610763611314 | 0.03778489101600335 | 0.03778489101600335 | 0.0001604756528216715 | 4.788117317683867e-05 |
| Go:0016192~vesicle-mediated transport | 2.6269187642578196 | 0.026155680242964663 | 0.3632422180079198 | 0.09571264878263674 | 0.2605320280071286 | 0.00022157919816047264 | 8.031139333139497e-05 |
| Go:0031175~neuron projection development | 3.64997107198942 | 0.01995706061474145 | 0.3893469659240534 | 0.07770429941719759 | 0.27642969307386667 | 0.000237532402557538 | 6.108568746053346e-05 |
| Go:0006644~phospholipid metabolic process | 4.13054772563975 | 0.071837560894916 | 0.5464434103913396 | 0.08688496865222228 | 0.3652138243452404 | 0.00033362059768355593 | 7.529869963277139e-05 |
| Go:0032313~regulation of rab gtpase activity | 9.416493416493417 | 0.22593681029350832 | 0.6533334174006811 | 0.08662576018402401 | 0.4193792487046456 | 0.00039908149623336275 | 3.69548392709718e-05 |
| Go:0032483~regulation of rab protein signal transduction | 9.416493416493417 | 0.22593681029350832 | 0.6533334174006811 | 0.08662576018402401 | 0.4193792487046456 | 0.00039908149623336275 | 3.69548392709718e-05 |
| Go:0000902~cell morphogenesis | 2.9712236508353014 | 0.05458185944186755 | 0.8745652160314754 | 0.09884698253606727 | 0.5173972539197562 | 0.0005347772461245756 | 0.00017007030177388404 |
| Go:0019637~organophosphate metabolic process | 3.8254504504504503 | 0.17872451230037845 | 0.9993633130917279 | 0.09889286014746634 | 0.5652781728655709 | 0.0006114495012186083 | 0.00014915302286524403 |
| Go:0005886~plasma membrane | 1.4560053390060272 | 0.07039792445344129 | 0.7965347746286211 | 0.07165303257331168 | 0.1381719080696714 | 0.0006193891722912183 | 0.0004146791113643423 |
| Mmu04722:neurotrophin signaling pathway | 4.619141323792487 | 0.02747634158291401 | 0.7035922751053914 | 0.01728932940129857 | 0.06738437535299457 | 0.0006226823108121126 | 0.0001254915683566321 |
| Mmu04510:focal adhesion | 3.70671834625323 | 0.017442583489065466 | 0.7060064763073415 | 0.013902921618077602 | 0.06760838263527535 | 0.000624825806490079 | 0.00015851444200885067 |
| Go:0032989~cellular component morphogenesis | 2.790072123405457 | 0.06848475893808126 | 1.0284474279421474 | 0.09087141122981113 | 0.5757441547605903 | 0.0006293309609410834 | 0.00021495485518416497 |
| Mmu04070:phosphatidylinositol signaling system | 6.227286821705427 | 0.05002774877429128 | 0.8885141353305648 | 0.014589105082495557 | 0.08440342860082373 | 0.0007870065367026434 | 0.0001155845449301496 |
| Mmu05220:chronic myeloid leukemia | 6.145348837209303 | 0.04102252621153446 | 0.9530899196951825 | 0.01342604960002558 | 0.09028048982152881 | 0.0008444553677562937 | 0.00012578095872042624 |
| Go:0006793~phosphorus metabolic process | 1.978985914320787 | 0.23947518518008237 | 1.3963849041395693 | 0.11008865856642014 | 0.6884932824590151 | 0.0008559729289850424 | 0.00041468147798482823 |
| Go:0006796~phosphate metabolic process | 1.978985914320787 | 0.23947518518008237 | 1.3963849041395693 | 0.11008865856642014 | 0.6884932824590151 | 0.0008559729289850424 | 0.00041468147798482823 |
| Head | 1.6367771168012566 | 0.12084327903193204 | 1.8232579988041175 | 0.05097986991337944 | 0.1888490182566882 | 0.0015724546874230773 | 0.0009261327630938868 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain | 1.7158772769473252 | 0.006535311703819824 | 0.08591089718871237 | 0.0060581281847249535 | 0.0060581281847249535 | 8.211221096744432e-05 | 4.716982526373326e-05 |
| Cell adhesion | 5.125550239234449 | 0.19696452027431133 | 6.2161593329033815 | 0.4495027516068164 | 0.4495027516068164 | 0.005723302616072238 | 0.0010746984800737687 |
| Alternative splicing | 1.6661946399951308 | 0.6261204993888361 | 8.870215169046725 | 0.3507753495349636 | 0.5785073532285512 | 0.008272830107863716 | 0.004811045180192287 |
| Cerebellum | 2.426693315567399 | 0.4838593744166445 | 11.892665042283578 | 0.3608301170284769 | 0.5914618607021696 | 0.012024018701674584 | 0.0047467990366617556 |
| Go:0007155~cell adhesion | 4.0368389780154486 | 0.6600600014619037 | 17.137148494819897 | 0.990516306780813 | 0.990516306780813 | 0.01372740483991105 | 0.0032643155046553603 |
| Go:0022610~biological adhesion | 4.029655990510083 | 0.6632987958665368 | 17.24872955238449 | 0.9042280922246919 | 0.9908277416810779 | 0.013825120859370972 | 0.0032932301600442954 |
| Mmu05211:renal cell carcinoma | 14.465546218487395 | 0.2073307842819464 | 14.573871485051459 | 0.595784128467725 | 0.595784128467725 | 0.01576573589448512 | 0.0010460708401495544 |
| Mmu04510:focal adhesion | 6.818775995246583 | 0.22542810471791797 | 14.99869064706727 | 0.37326858361022275 | 0.6072077317100637 | 0.016260632536822837 | 0.0022986584924110046 |
| Corpora quadrigemina | 3.7547009354625343 | 0.5093299520506558 | 19.1400817377681 | 0.39388276619897766 | 0.7773258015957688 | 0.02009329722709151 | 0.0051090865621192935 |
| Go:0005794~golgi apparatus | 3.130248193977541 | 0.6641663197100003 | 20.40445033672631 | 0.876240829425298 | 0.876240829425298 | 0.020276106153103678 | 0.0061804765702969285 |
| Mmu04012:erbb signaling pathway | 11.638945233265721 | 0.4802664589852198 | 21.207324451926524 | 0.3667414288405725 | 0.7460529161594871 | 0.023759324492495845 | 0.0019633524390691715 |
| Golgi apparatus | 3.522077922077922 | 0.7557603695612728 | 25.064319365925513 | 0.5912273657217841 | 0.9316961094776957 | 0.025475538882615768 | 0.006886012617688139 |
| Go:0030017~sarcomere | 11.436585365853658 | 0.5485277089542041 | 26.240778887572315 | 0.7517498202942179 | 0.9383718482760469 | 0.026950150894546485 | 0.0022566580886310074 |
| Go:0044449~contractile fiber part | 10.638684061259216 | 0.3865908313736287 | 29.428111183136764 | 0.6548210793043174 | 0.9588724537538046 | 0.030800756098812127 | 0.0027729838408344603 |
| Go:0016192~vesicle-mediated transport | 4.049833094897473 | 0.9102283761540358 | 34.73204036244364 | 0.9705261215247989 | 0.9999743957614021 | 0.030886122410274563 | 0.007277891027930105 |
| Go:0030016~myofibril | 10.05414098097025 | 0.4365953529435477 | 32.110797553449956 | 0.5878942770635693 | 0.9711573883437026 | 0.034166422229574285 | 0.0032546759726419876 |
| Go:0043292~contractile fiber | 9.630808729139922 | 0.856852690947361 | 34.26504604660304 | 0.5361652489628899 | 0.9785307725600825 | 0.036957931070074494 | 0.0036748241356467774 |
| Immunoglobulin domain | 3.6638620972706755 | 0.9067744051360538 | 40.04693972185378 | 0.6956727122481656 | 0.9914224436094125 | 0.04472481572224427 | 0.01156366487109709 |
| Go:0032982~myosin filament | 40.66341463414635 | 0.8171517204113358 | 41.47645198158065 | 0.5584680991577247 | 0.992590779461999 | 0.046950589895166164 | 0.001072088124232418 |
| Mmu04722:neurotrophin signaling pathway | 7.789140271493213 | 0.7442926487685522 | 39.656387090107714 | 0.5162390521674092 | 0.9452324610169374 | 0.0496822774610645 | 0.0061240613286317434 |
| Go:0005793~er-golgi intermediate compartment | 38.1219512195122 | 0.5773914674544844 | 43.53108966714313 | 0.5264335860447937 | 0.9946584627924179 | 0.050003034158402127 | 0.001222702150376853 |
| Mmu04910:insulin signaling pathway | 7.337595907928389 | 0.5530453087823217 | 43.096519834067436 | 0.4771395282935236 | 0.9609222948370462 | 0.055293288118850405 | 0.007228683869581828 |
| Mmu05200:pathways in cancer | 4.179930795847751 | 0.7681104024877493 | 44.31819593717672 | 0.42945757489464864 | 0.9655072625571148 | 0.057359490645411 | 0.013080310998412543 |
| Go:0032989~cellular component morphogenesis | 4.301361190250079 | 0.9884955704255679 | 57.45454045214346 | 0.9949792544973158 | 0.9999999993645625 | 0.06090524503540353 | 0.013439249088701716 |
| Mmu04310:wnt signaling pathway | 6.795894196604817 | 0.6305861733536393 | 47.73507288100497 | 0.4131726708681065 | 0.976035282259904 | 0.06336271004337843 | 0.008931387619385169 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0043009~chordate embryonic development | 16.137767220902614 | 0.973972423074804 | 61.56209570570705 | 0.9973913126503394 | 0.9973913126503394 | 0.09010594828999482 | 0.005512927508769393 |
| Go:0009792~embryonic development ending in birth or egg hatching | 15.985882352941175 | 0.8440975640128066 | 61.91525348405891 | 0.9503704439371569 | 0.9975369071650051 | 0.09093500888081586 | 0.005616064186674952 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain cortex | 2.1776440128682895 | 1.7447863869575083e-24 | 4.439991477971947e-23 | 9.231619566410946e-24 | 9.231619566410946e-24 | 3.3692042213178634e-26 | 1.2552420050054018e-26 |
| Brain | 1.242052474142542 | 1.1830027100166298e-23 | 1.4496742460211878e-22 | 1.507079596986536e-23 | 3.014159193973072e-23 | 1.1000580999901721e-25 | 8.510810863428991e-26 |
| Membrane | 1.2727529723393254 | 5.475152153975962e-22 | 5.289543976936313e-21 | 2.1272216766064966e-21 | 2.1272216766064966e-21 | 3.5872203652723384e-24 | 2.683898114694099e-24 |
| Phosphoprotein | 1.199711824961826 | 2.2648549702353193e-14 | 3.219646771412954e-13 | 6.583622536027178e-14 | 1.3167245072054357e-13 | 1.7290111474222422e-16 | 1.6079777408986103e-16 |
| Hippocampus | 1.6559573641001188 | 3.086420008457935e-14 | 2.886579864025407e-13 | 2.0317081350640365e-14 | 6.084022174945858e-14 | 2.2531093362033743e-16 | 1.7278596915463814e-16 |
| Medulla oblongata | 1.893865694149467 | 2.3148150063434514e-12 | 4.916067553040193e-11 | 2.5552893134772603e-12 | 1.0221157253909041e-11 | 3.7289630193568015e-14 | 1.6610627858547365e-14 |
| Liver | 1.2778923440339822 | 1.772839652858238e-10 | 2.2679080835530385e-09 | 9.430844993829623e-11 | 4.715421386691787e-10 | 1.7209924858436352e-12 | 1.2754431672415437e-12 |
| Go:0044456~synapse part | 2.260054023115467 | 6.229718962913466e-10 | 1.5106293993483177e-08 | 5.782414547184089e-09 | 5.782414547184089e-09 | 1.0325697734090817e-11 | 3.5396493738316843e-12 |
| Go:0006812~cation transport | 1.7553358001572226 | 1.0003941008918105e-08 | 1.1379021058743888e-07 | 2.2974495361705038e-07 | 2.2974495361705038e-07 | 6.170968091386817e-11 | 3.0314982988716304e-11 |
| Olfactory bulb | 2.2338834678552786 | 3.2239971803349476e-09 | 8.568702414279983e-08 | 2.9693374425932006e-09 | 1.78160245445369e-08 | 6.502203228656956e-11 | 2.319420493557936e-11 |
| Eye | 1.4198141786135532 | 6.8575159994921364e-09 | 9.968275094252022e-08 | 2.9608588913987433e-09 | 2.0726012239791203e-08 | 7.564232823035511e-11 | 4.933461211925169e-11 |
| Go:0045202~synapse | 1.9400568233953563 | 9.292660863025048e-09 | 1.808245331247349e-07 | 3.460816022649027e-08 | 6.921631912071291e-08 | 1.236005766754242e-10 | 5.2799154238588077e-11 |
| Go:0006811~ion transport | 1.6042991000805467 | 5.097220179273165e-08 | 5.148773074559188e-07 | 5.197742652951476e-07 | 1.0395482604730333e-06 | 2.792234267491813e-10 | 1.5446121917002689e-10 |
| Potassium | 2.609686501051775 | 8.828021524909246e-08 | 7.729752282692459e-07 | 1.036188617131728e-07 | 3.108565528320284e-07 | 5.242101740800672e-10 | 1.4424872876017426e-10 |
| Kinase | 1.581282891265183 | 7.040668337232603e-08 | 8.961548281227749e-07 | 9.009849977203999e-08 | 3.6039395057141377e-07 | 6.07747027697869e-10 | 3.4513082521615727e-10 |
| Go:0030001~metal ion transport | 1.7757158959112667 | 9.996448169236061e-08 | 1.1566768320925291e-06 | 7.784518420983488e-07 | 2.3353537083048437e-06 | 6.272781397594587e-10 | 3.029226673132874e-10 |
| Potassium transport | 2.6902782012061577 | 3.701810258327498e-08 | 1.0210939760391113e-06 | 8.212780411298581e-08 | 4.106389533964361e-07 | 6.924773298722696e-10 | 1.8146124493066593e-10 |
| Go:0048878~chemical homeostasis | 1.8623298268419677 | 1.0440635811459487e-07 | 1.2836590568277018e-06 | 6.479338841014481e-07 | 2.5917330175317943e-06 | 6.961419379078851e-10 | 3.1638288674731407e-10 |
| Synapse | 2.1371835562808172 | 1.1174911573874624e-07 | 1.0840969122405397e-06 | 7.266267121330827e-08 | 4.35975948120948e-07 | 7.35204133119996e-10 | 2.7389488318617833e-10 |
| Corpora quadrigemina | 1.688733398692853 | 6.925409290481355e-08 | 1.2602287213780983e-06 | 3.2753306289379225e-08 | 2.620264198949229e-07 | 9.563009719780085e-10 | 4.982308737107835e-10 |
| Cytoplasm | 1.2420799661514743 | 2.5993888874698e-07 | 2.3684691652370304e-06 | 1.3607056481568236e-07 | 9.524935645766064e-07 | 1.6062293359915974e-09 | 1.274210448307257e-09 |
| Go:0006873~cellular ion homeostasis | 2.0137172259666967 | 7.844076918006593e-07 | 3.563011208296274e-06 | 1.438758866934009e-06 | 7.193773634339706e-06 | 1.932258774106796e-09 | 7.923313394744221e-10 |
| Kidney | 1.3106183301554861 | 1.9592868538698127e-07 | 2.5656839985899182e-06 | 5.9272932007026213e-08 | 5.334562614978111e-07 | 1.946921125499327e-09 | 1.409559056519177e-09 |
| Go:0030425~dendrite | 2.601345009434419 | 1.0241861059245849e-07 | 3.0856490140784842e-06 | 3.9370990601028666e-07 | 1.1811292530694573e-06 | 2.1091605746472815e-09 | 5.819239051720818e-10 |
| Go:0005783~endoplasmic reticulum | 1.500859264753887 | 2.4181123525757187e-07 | 3.295719397389263e-06 | 3.1538521294560695e-07 | 1.261540255037552e-06 | 2.2527519182521694e-09 | 1.3739276206529753e-09 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain cortex | 2.348247068213913 | 6.322125581268313e-23 | 1.6530081063015303e-21 | 3.181074301505522e-22 | 3.181074301505522e-22 | 1.2724297206022087e-24 | 4.548291785085117e-25 |
| Brain | 1.2719127755149493 | 4.016608012758439e-22 | 4.935852999840725e-21 | 4.749315830317576e-22 | 9.498631660635152e-22 | 3.799452664254061e-24 | 2.8896460523442006e-24 |
| Membrane | 1.273608669232078 | 1.8468607812807283e-16 | 1.7466592200173581e-15 | 6.624122242768336e-16 | 6.624122242768336e-16 | 1.1956899355177502e-18 | 9.053239123925138e-19 |
| Hippocampus | 1.7846919847355267 | 3.079991828714088e-15 | 5.667338875256363e-14 | 3.635438121421336e-15 | 1.090631436426401e-14 | 4.3625257457056036e-17 | 2.215821459506538e-17 |
| Phosphoprotein | 1.2276916913565115 | 1.1324274851176597e-13 | 9.769962616701378e-13 | 1.8451906669270102e-13 | 3.6903813338540203e-13 | 6.662369474100327e-16 | 5.086413391030823e-16 |
| Medulla oblongata | 2.0826883852489892 | 5.864198016070077e-13 | 1.3411494137471891e-11 | 6.45261621912141e-13 | 2.581268532253489e-12 | 1.0277636218209605e-14 | 4.247551597179677e-15 |
| Eye | 1.541992698873412 | 8.38580316298021e-11 | 1.3346879157438707e-09 | 5.137001934940599e-11 | 2.5685009674702997e-10 | 1.0274354594613675e-12 | 6.033439120964011e-13 |
| Synapse | 2.51424236436195 | 1.122824500043862e-09 | 1.2648981861929087e-08 | 1.5990216928329914e-09 | 4.797065189521277e-09 | 8.658968377804742e-12 | 2.7519676373396537e-12 |
| Go:0045202~synapse | 2.1746553871258643 | 8.83419559727372e-10 | 1.863406096092035e-08 | 6.314029299403501e-09 | 6.314029299403501e-09 | 1.2991801601396408e-11 | 5.019436518012085e-12 |
| Go:0044456~synapse part | 2.423882140087878 | 4.835076339304578e-09 | 1.1815094458356157e-07 | 2.001733956369378e-08 | 4.0034678794320655e-08 | 8.2375922447538e-11 | 2.747199996725902e-11 |
| Go:0048878~chemical homeostasis | 2.0626662033396124 | 2.1274545236593667e-08 | 2.813386079658642e-07 | 5.210352399531715e-07 | 5.210352399531715e-07 | 1.5419807455030755e-10 | 6.446833661675639e-11 |
| Visual cortex | 2.3654671961771925 | 2.584040303954538e-08 | 6.876567537617007e-07 | 2.2055620996219716e-08 | 1.3233371853882403e-07 | 5.29334933495276e-10 | 1.8590218388683538e-10 |
| Go:0019226~transmission of nerve impulse | 2.3278944196982407 | 1.3776023710665442e-07 | 2.134880738857703e-06 | 1.9768835262112816e-06 | 3.953763144326494e-06 | 1.1701009337382634e-09 | 4.174552698422891e-10 |
| Liver | 1.2725527872767737 | 2.3330343368321849e-07 | 2.880304805774614e-06 | 7.9184326651216e-08 | 5.542901551081059e-07 | 2.2171612549190833e-09 | 1.6784421737473902e-09 |
| Go:0006873~cellular ion homeostasis | 2.189494038027715 | 1.091306770839573e-06 | 5.20154236349768e-06 | 3.211057504337589e-06 | 9.633141580311921e-06 | 2.8508991112108767e-09 | 1.1023306940248965e-09 |
| Go:0055082~cellular chemical homeostasis | 2.1661518841041785 | 4.2730246230249236e-07 | 6.025537391263214e-06 | 2.7898001003778816e-06 | 1.1159153703865776e-05 | 3.3025202215363544e-09 | 1.294856220404287e-09 |
| Cerebellum | 1.4246598397572654 | 5.124570879866397e-07 | 7.186046158125237e-06 | 1.728617173846203e-07 | 1.3828929026349357e-06 | 5.531575460569296e-09 | 3.68674255832908e-09 |
| Go:0050801~ion homeostasis | 2.0742013241774018 | 1.1974030946770853e-06 | 1.596944886328444e-05 | 5.915024635760879e-06 | 2.957477330567393e-05 | 8.752651858160476e-09 | 3.6284963501944554e-09 |
| Go:0030001~metal ion transport | 1.8469875369270643 | 1.4099892668983927e-06 | 1.607280774029718e-05 | 4.961092682886914e-06 | 2.9766186913304793e-05 | 8.809301546468148e-09 | 4.272697796451721e-09 |
| Alternative splicing | 1.2066864696794841 | 1.573904964802253e-06 | 1.3998226500167021e-05 | 1.3271894627742142e-06 | 5.308747282550819e-06 | 9.582601726149915e-09 | 7.715226464400455e-09 |
| Olfactory bulb | 2.273699542238868 | 4.975972330845124e-07 | 1.2581209640760704e-05 | 2.690163576346549e-07 | 2.4211446132405e-06 | 9.684590171292325e-09 | 3.5798371367571633e-09 |
| Corpora quadrigemina | 1.7534242430355234 | 8.149258026168837e-07 | 1.4812049897727775e-05 | 2.850453538760789e-07 | 2.850449882130235e-06 | 1.140181576400815e-08 | 5.862777915990852e-09 |
| Go:0005886~plasma membrane | 1.2584280044122222 | 1.735947891323697e-06 | 1.7947634689541303e-05 | 2.0271445106967434e-06 | 6.08142120395172e-06 | 1.2513250317792827e-08 | 9.86334880011821e-09 |
| Go:0006812~cation transport | 1.7613128183856792 | 2.702792205599991e-06 | 2.854445632083369e-05 | 7.551966265451071e-06 | 5.286256619685403e-05 | 1.564485439163145e-08 | 8.190290424656531e-09 |
| Kinase | 1.6126850918556501 | 2.627460410353777e-06 | 3.356704048051995e-05 | 2.5460253630926744e-06 | 1.2730061993315722e-05 | 2.2978597202756782e-08 | 1.2879724701653157e-08 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain cortex | 2.66056718287652 | 1.9204063233013478e-22 | 5.4482740933906355e-21 | 9.738554454074817e-22 | 9.738554454074817e-22 | 4.2526438664082176e-24 | 1.38158728295061e-24 |
| Brain | 1.3279774841020746 | 6.307521813981239e-22 | 7.934840631079718e-21 | 7.091592332323833e-22 | 1.4183184664647665e-21 | 6.193530421243522e-24 | 4.537785477684344e-24 |
| Membrane | 1.3330167199787681 | 1.731282440532801e-16 | 1.6780526784157973e-15 | 5.806370326913223e-16 | 5.806370326913223e-16 | 1.1659378166492416e-18 | 8.48667863006275e-19 |
| Hippocampus | 1.966952337544924 | 3.330339099407769e-15 | 6.555656173004098e-14 | 3.905984019198566e-15 | 1.1717952057595698e-14 | 5.1170096321378596e-17 | 2.395927409645877e-17 |
| Medulla oblongata | 2.3096954196698443 | 2.3456792064280307e-12 | 5.575540029667536e-11 | 2.4915625118637763e-12 | 9.966250047455105e-12 | 4.355147192196492e-14 | 1.6882342954525988e-14 |
| Eye | 1.6904961149736233 | 1.2237655333535713e-11 | 2.0226043062621102e-10 | 7.2306605147787195e-12 | 3.615308052928867e-11 | 1.5791680190777477e-13 | 8.803702444705058e-14 |
| Synapse | 3.0241964608161793 | 6.074341030171126e-11 | 7.931766354829506e-10 | 1.37227562646558e-10 | 2.74455125293116e-10 | 5.510596678644594e-13 | 1.4883655531940316e-13 |
| Go:0045202~synapse | 2.545777905524966 | 5.760369958807132e-11 | 1.3837597734323026e-09 | 4.246738516400228e-10 | 4.246738516400228e-10 | 9.807293364234002e-13 | 3.273378040065889e-13 |
| Phosphoprotein | 1.242153504869398 | 1.9833334974350691e-10 | 1.8078205599181274e-09 | 2.0851365079010975e-10 | 6.255409523703293e-10 | 1.2561306059627197e-12 | 9.722197212104078e-13 |
| Go:0044456~synapse part | 2.9293398929556145 | 1.333990695684406e-10 | 3.814193405560218e-09 | 5.852845985643285e-10 | 1.170569197128657e-09 | 2.7034263354383596e-12 | 7.579919221756394e-13 |
| Go:0019226~transmission of nerve impulse | 2.833359745773315 | 3.3172198321551605e-09 | 6.073505032233584e-08 | 9.806473910689562e-08 | 9.806473910689562e-08 | 3.386213000360979e-11 | 1.0052233560943836e-11 |
| Visual cortex | 2.7643337422197996 | 7.137824664660286e-09 | 2.140526866867276e-07 | 6.376832173415892e-09 | 3.826099248538384e-08 | 1.6707861722872087e-10 | 5.135129987556435e-11 |
| Corpora quadrigemina | 1.9973129152740423 | 7.939004975643371e-08 | 1.5934993879973547e-06 | 4.069016057428598e-08 | 2.848310892700212e-07 | 1.2438040763453396e-09 | 5.711514557069127e-10 |
| Cerebellum | 1.5407715014759593 | 1.3342290339224405e-07 | 1.9709325393968413e-06 | 4.40369586351963e-08 | 3.522956144585976e-07 | 1.5384091163057471e-09 | 9.598770166702311e-10 |
| Go:0006873~cellular ion homeostasis | 2.502475435232431 | 7.148625242869855e-07 | 3.739754039777665e-06 | 3.0191583261229837e-06 | 6.038307536870846e-06 | 2.0850572234714407e-09 | 7.220836441020601e-10 |
| Go:0055082~cellular chemical homeostasis | 2.437112270879345 | 6.229331848128794e-07 | 9.467280570607528e-06 | 5.095377897390563e-06 | 1.5286055803587217e-05 | 5.278374528458632e-09 | 1.8876768497885376e-09 |
| Go:0050801~ion homeostasis | 2.3602950400967595 | 6.892473488617767e-07 | 1.0063236255408725e-05 | 4.0620971766269776e-06 | 1.6248289703035823e-05 | 5.610642842171599e-09 | 2.088629051562912e-09 |
| Go:0048878~chemical homeostasis | 2.1753992743986674 | 1.2622911866877473e-06 | 1.6703669858397774e-05 | 5.394041082240619e-06 | 2.6969914456054234e-05 | 9.312941344062141e-09 | 3.825127200846104e-09 |
| Go:0005886~plasma membrane | 1.3027645598376643 | 4.693047884196666e-06 | 5.029756059427015e-05 | 5.1454061957834085e-06 | 1.543613916188402e-05 | 3.564955662624668e-08 | 2.6665106977754213e-08 |
| Cell junction | 2.0463108320251178 | 3.2794585786977137e-06 | 5.328701535578517e-05 | 4.609561995438938e-06 | 1.8438120493846633e-05 | 3.7024679044679045e-08 | 1.607580355236175e-08 |
| Cell membrane | 1.440298172347209 | 6.085724358340094e-06 | 6.451076900448882e-05 | 4.464373549506462e-06 | 2.2321668442182308e-05 | 4.482312666782584e-08 | 2.9832072496445006e-08 |
| Go:0043005~neuron projection | 2.3397929945617215 | 3.3740688284433773e-06 | 7.275665131478704e-05 | 5.582216318855693e-06 | 2.232867830931351e-05 | 5.156796080892648e-08 | 1.9170877825047983e-08 |
| Go:0019725~cellular homeostasis | 2.1282414137056462 | 1.202084590901098e-05 | 0.00015517436934286977 | 4.175740477496692e-05 | 0.0002505182748929746 | 8.651576281010529e-08 | 3.642702400931863e-08 |
| Go:0007242~intracellular signaling cascade | 1.609596061041496 | 4.628506020321055e-05 | 0.00043789634369684194 | 0.00010100102665144828 | 0.0007067929972631726 | 2.4414461568099593e-07 | 1.4026099437352667e-07 |
| Go:0042391~regulation of membrane potential | 3.01336142374009 | 0.0001522923534439391 | 0.0006045200400150996 | 0.0001220025830623861 | 0.0009756039965301833 | 3.370442139187289e-07 | 9.230542066629224e-08 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Secreted | 3.5088110055682935 | 0.018555429834166026 | 0.37758234323970186 | 0.038254291622277226 | 0.038254291622277226 | 0.00033049791306361497 | 9.18082435755705e-05 |
| Go:0005576~extracellular region | 2.7064935064935063 | 0.1265812558348598 | 2.22548144681054 | 0.1485669731042688 | 0.1485669731042688 | 0.002142161424392996 | 0.0007686826904680446 |
| Signal | 2.2368177903061626 | 0.17640896275334794 | 2.4763712788285708 | 0.12126629024919433 | 0.22782706734758684 | 0.0021886754849928273 | 0.0009509259472959223 |
| Go:0044421~extracellular region part | 3.916373032652103 | 0.22434804227004101 | 3.02128369571677 | 0.10382392084275072 | 0.19686843514633956 | 0.0029188882286126767 | 0.0007214764500718595 |
| Pyrrolidone carboxylic acid | 29.657807308970096 | 0.10789996766949028 | 4.90198579197415 | 0.1586490374789491 | 0.40443168081847014 | 0.004382227279079738 | 0.00013991301618918473 |
| Go:0005584~collagen type i | 378.90909090909093 | 0.08747416110203887 | 5.2367428489368155 | 0.12025940946245406 | 0.3191304824263992 | 0.005112016877878618 | 6.754616895728707e-06 |
| Go:0001944~vasculature development | 8.052148148148147 | 0.6051454273349854 | 14.902916021520351 | 0.98470088653265 | 0.98470088653265 | 0.011705487743433775 | 0.0014069453115124167 |
| Mammary tumor. wap-tgf alpha model. 7 months old | 5.312299465240641 | 0.2831208761695204 | 12.206067615927907 | 0.5120580285804772 | 0.5120580285804772 | 0.013200228381502987 | 0.0023917254617517227 |
| Extracellular matrix | 7.7973577901517634 | 0.4981369805438397 | 14.56039727853431 | 0.33344025442199354 | 0.8025958266103413 | 0.013655917337707019 | 0.0016883478064863358 |
| Go:0005583~fibrillar collagen | 108.25974025974025 | 0.5999494985795062 | 17.163153951394094 | 0.28566545589769243 | 0.7396206072953542 | 0.017781548945420736 | 0.00014067875343096238 |
| Triple helix | 92.26873385012921 | 0.5033684750441625 | 21.544391008591656 | 0.39368377087683515 | 0.9180599989388544 | 0.02097825345234094 | 0.00020179824220511086 |
| Disulfide bond | 2.0180282008533723 | 0.9072043062034525 | 24.686694969835944 | 0.385659049206185 | 0.9462403053689102 | 0.024468810208351274 | 0.011586059829393973 |
| Go:0048705~skeletal system morphogenesis | 11.613675213675213 | 0.9925020457614535 | 29.726798457271684 | 0.9896296909350989 | 0.9998924566898985 | 0.025411322215340707 | 0.0021159940798073673 |
| Leucine-rich repeat | 6.039408033826638 | 0.5796563935115668 | 26.6169898889839 | 0.3660904535697357 | 0.9588669375536386 | 0.026679531903749155 | 0.00423943318028644 |
| Glycoprotein | 1.7300387596899225 | 0.9724402507049208 | 30.71988158663277 | 0.37688283290068725 | 0.9772721826220949 | 0.03156043682845229 | 0.017450835919024788 |
| Go:0015031~protein transport | 3.8652784889344027 | 0.9370071987561841 | 37.51875891115583 | 0.9827607543392344 | 0.9999948766411553 | 0.033732578045314 | 0.008342986812220354 |
| Go:0045184~establishment of protein localization | 3.8358175248419153 | 0.9423694708694402 | 38.24707356759768 | 0.9559023883112951 | 0.9999962185333421 | 0.034558854932537394 | 0.00861030152828313 |
| Citrullination | 51.901162790697676 | 0.5572886182075214 | 35.04268074319945 | 0.38998604629535094 | 0.9883034461623642 | 0.03699706771475272 | 0.0006654951506815841 |
| Go:0005578~proteinaceous extracellular matrix | 5.103152739516376 | 0.730322357518008 | 34.57532378118492 | 0.4546826127845137 | 0.9517778326350785 | 0.03961959659795961 | 0.007418527406180779 |
| Developmental protein | 3.0605108279526885 | 0.9267964918469304 | 38.141148026800906 | 0.3905753998295265 | 0.9929335740759843 | 0.04110096901552883 | 0.012734456764818871 |
| Go:0031012~extracellular matrix | 4.904972050603119 | 0.7777298786756921 | 37.45552585376869 | 0.4281881159855998 | 0.9650442147531717 | 0.04373061032583184 | 0.008508267803035059 |
| Go:0005581~collagen | 39.88516746411483 | 0.7944639282779963 | 40.03075180800521 | 0.4066793120934248 | 0.9741162950233885 | 0.04755402123478161 | 0.0011230434375893137 |
| Go:0008104~protein localization | 3.341694948600659 | 0.9933040005680267 | 52.577555650902205 | 0.9790356104845207 | 0.9999999959504096 | 0.05298053730258075 | 0.015055948062559096 |
| Go:0043588~skin development | 33.550617283950615 | 0.9999999727631352 | 54.54049861153292 | 0.9667356581226143 | 0.999999998645205 | 0.05589703340660609 | 0.001598234267272194 |
| Go:0006955~immune response | 4.273963985216638 | 0.9878975499908537 | 56.956411460684585 | 0.9558054328869506 | 0.9999999996707072 | 0.05965122331044131 | 0.013287738285154614 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| None |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| None |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain cortex | 1.9996264326127955 | 2.084844224320196e-25 | 5.130763617643755e-24 | 1.1388302862078067e-24 | 1.1388302862078067e-24 | 3.8473996155669144e-27 | 1.4998879311656084e-27 |
| Brain | 1.2036290688542286 | 3.555272866176445e-23 | 4.2998397854740845e-22 | 4.7719873087701916e-23 | 9.543974617540383e-23 | 3.224315749169048e-25 | 2.5577502634362915e-25 |
| Phosphoprotein | 1.1977087552013588 | 6.167896711202742e-20 | 5.643253018046539e-19 | 2.3616922315293287e-19 | 2.3616922315293287e-19 | 3.8030470717058432e-22 | 3.023478780001344e-22 |
| Mammary tumor | 1.3627606655986357 | 1.7070450183780902e-18 | 2.426736473214592e-17 | 1.7954708748344613e-18 | 5.386412624503384e-18 | 1.8197339947646567e-20 | 1.2280899412792016e-20 |
| Liver | 1.3011539694754393 | 1.1755038962526964e-17 | 1.5722998803363625e-16 | 8.724737954066304e-18 | 3.4898951816265215e-17 | 1.1790186424413924e-19 | 8.456862562969039e-20 |
| Membrane | 1.2013472259604048 | 8.455248368138072e-17 | 7.766502882825858e-16 | 1.6251344274183579e-16 | 3.2502688548367157e-16 | 5.233927302474583e-19 | 4.144729592224545e-19 |
| Kidney | 1.3919321533923306 | 4.7800802115934685e-17 | 6.989376578961495e-16 | 3.102740380578387e-17 | 1.551370190289193e-16 | 5.241115507733761e-19 | 3.438906627045661e-19 |
| Go:0005739~mitochondrion | 1.4816717063962037 | 2.3887230711073284e-16 | 3.3496053095148565e-15 | 1.3509834018381734e-15 | 1.3509834018381734e-15 | 2.270560339223821e-18 | 1.3572290176746184e-18 |
| Cytoplasm | 1.289178583300578 | 2.1116577782147416e-15 | 2.1286877201683458e-14 | 2.9695078565481395e-15 | 8.908523569644419e-15 | 1.4345448582358163e-17 | 1.0351263618699714e-17 |
| Hippocampus | 1.5475914649447111 | 3.086420008457935e-14 | 4.440892098500626e-13 | 1.6431300764452317e-14 | 9.85878045867139e-14 | 3.784755580695146e-16 | 1.998986033655157e-16 |
| Medulla oblongata | 1.7619834259710165 | 7.716050021144838e-13 | 1.6142642778049776e-11 | 5.117017920497346e-13 | 3.582023566650605e-12 | 1.2115496691226844e-14 | 5.6034331391830105e-15 |
| Visual cortex | 2.2025242218492522 | 5.70987701564718e-13 | 1.6431300764452317e-11 | 4.559685962135518e-13 | 3.647748769708414e-12 | 1.2314322583116208e-14 | 4.117964212465332e-15 |
| Alternative splicing | 1.2030901745884077 | 3.148148408627094e-12 | 2.915445662665661e-11 | 3.0507818493674677e-12 | 1.2203238419772333e-11 | 1.96407308208973e-14 | 1.541749342544996e-14 |
| Mitochondrion | 1.5243972445464982 | 2.37583286377685e-10 | 3.046429775110937e-09 | 2.549855881994745e-10 | 1.2749281630419773e-09 | 2.0530144010475197e-12 | 1.164596034827628e-12 |
| Colon | 1.4553488016578058 | 9.357253860642345e-10 | 1.500706225954218e-08 | 3.7010960962646777e-10 | 3.3309861535713026e-09 | 1.1253292787644903e-11 | 6.7318524197893684e-12 |
| Go:0045202~synapse | 1.8193614265466445 | 2.307450230887298e-09 | 4.3991332798754e-08 | 8.871427481516037e-09 | 1.774285485200977e-08 | 2.9819936854568726e-11 | 1.3110460200796292e-11 |
| Go:0031090~organelle membrane | 1.479636722593984 | 5.272907444009434e-09 | 7.381553146501574e-08 | 9.923911803433327e-09 | 2.9771735188255377e-08 | 5.0036496082240076e-11 | 2.9959657856311233e-11 |
| Eye | 1.3461814925232434 | 1.813172978426536e-08 | 2.5448024798535585e-07 | 5.648473244690422e-09 | 5.6484730892591983e-08 | 1.908267997499492e-10 | 1.3044404792009603e-10 |
| Go:0044456~synapse part | 1.9554457381010097 | 4.027518507587757e-08 | 8.595412270828717e-07 | 8.66688593115228e-08 | 3.466753922820587e-07 | 5.826478502636312e-10 | 2.2883630404349175e-10 |
| Go:0005773~vacuole | 1.9559250140172109 | 8.636193260525005e-08 | 1.8441311810946104e-06 | 1.4875725451446442e-07 | 7.437860514158956e-07 | 1.2500610138574725e-09 | 4.906927990612852e-10 |
| Synapse | 1.9247209240902747 | 2.3816722838354565e-07 | 2.1600079946892947e-06 | 1.5065996372687351e-07 | 9.03959442077884e-07 | 1.4556519064619033e-09 | 5.837433181751018e-10 |
| Mmu04142:lysosome | 2.340703271599902 | 1.2239357249921312e-07 | 2.262036158384717e-06 | 3.412416278436581e-07 | 3.412416278436581e-07 | 1.824821847596158e-09 | 5.513224637023584e-10 |
| Go:0008021~synaptic vesicle | 2.743375344335828 | 9.76236501637473e-08 | 3.7897576410372835e-06 | 2.547513630046794e-07 | 1.5285072042514614e-06 | 2.5689216756858212e-09 | 5.546798604741982e-10 |
| Go:0005764~lysosome | 2.008726141639155 | 1.8270465440561878e-07 | 4.071143844264213e-06 | 2.3457122855230494e-07 | 1.6419974444570329e-06 | 2.7596618017829205e-09 | 1.0380947175764349e-09 |
| Go:0000323~lytic vacuole | 1.997504207886981 | 2.4202064463629824e-07 | 5.345015008373366e-06 | 2.694730089070063e-07 | 2.1557820384376924e-06 | 3.623167043380246e-09 | 1.3751174510033975e-09 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain cortex | 2.12490756766392 | 2.3514001309783058e-23 | 5.813809171754282e-22 | 1.1976125677381071e-22 | 1.1976125677381071e-22 | 4.419234567299289e-25 | 1.6916547704879898e-25 |
| Brain | 1.2321395142863598 | 1.4366843596148764e-22 | 1.743016434624289e-21 | 1.7952587763129462e-22 | 3.5905175526258925e-22 | 1.324914226061215e-24 | 1.0335858702265298e-24 |
| Phosphoprotein | 1.2090656341528614 | 1.0075258731422739e-16 | 9.148772184384616e-16 | 3.668523518050813e-16 | 3.668523518050813e-16 | 6.207315597378702e-19 | 4.938852319324872e-19 |
| Kidney | 1.4257969577119542 | 1.5432100042289676e-14 | 2.886579864025407e-13 | 2.0095036745715333e-14 | 6.017408793468348e-14 | 1.7520029495979818e-16 | 1.5284498477765016e-16 |
| Membrane | 1.2160651413616443 | 6.794564910705958e-14 | 4.884981308350689e-13 | 9.836575998178887e-14 | 1.9684254226604025e-13 | 3.2934231921647965e-16 | 3.068144553808063e-16 |
| Hippocampus | 1.6401010755354635 | 3.086420008457935e-14 | 4.3298697960381105e-13 | 2.2537527399890678e-14 | 9.026113190202523e-14 | 3.8068429471507293e-16 | 1.974765274096502e-16 |
| Medulla oblongata | 1.9438540490137284 | 3.086420008457935e-14 | 7.327471962526033e-13 | 3.008704396734174e-14 | 1.504352198367087e-13 | 5.143043578029251e-16 | 2.3792339223933187e-16 |
| Liver | 1.3089192198008328 | 1.851852005074761e-13 | 2.3314683517128287e-12 | 8.026912468039882e-14 | 4.813927034774679e-13 | 1.8237626786089524e-15 | 1.3248011572763145e-15 |
| Visual cortex | 2.4631042949237414 | 1.0802470029602773e-13 | 3.3639757646142243e-12 | 9.880984919163893e-14 | 6.920020112488601e-13 | 2.5858802798282453e-15 | 7.800316722321831e-16 |
| Mammary tumor | 1.3507426363746413 | 1.4043211038483605e-12 | 1.942890293094024e-11 | 5.00155472593633e-13 | 4.001576847656452e-12 | 1.4762922180438322e-14 | 1.0074744580383674e-14 |
| Eye | 1.4568560754490332 | 6.402778307545987e-11 | 9.505396469933203e-10 | 2.1756263457461955e-11 | 1.9580648213946006e-10 | 7.225252690758008e-13 | 4.605945613391319e-13 |
| Alternative splicing | 1.210938559757757 | 9.031335679310359e-10 | 7.863842910182939e-09 | 1.0510954329134847e-09 | 3.153286298740454e-09 | 5.335455582173812e-12 | 4.427180446765296e-12 |
| Cytoplasm | 1.2713313783750542 | 1.112179681683756e-09 | 1.0884992907023161e-08 | 1.091180257262181e-09 | 4.364720918026421e-09 | 7.385300854438228e-12 | 5.451805892170354e-12 |
| Go:0005739~mitochondrion | 1.420491762397965 | 5.060801555600847e-09 | 6.738474223055846e-08 | 2.5833331807056936e-08 | 2.5833331807056936e-08 | 4.6048693389928915e-11 | 2.8754514701695994e-11 |
| Colon | 1.4980876831444805 | 1.3782254115746184e-08 | 2.0862609417804379e-07 | 4.297582179013659e-09 | 4.297582123502508e-08 | 1.5858233526565034e-10 | 9.91529352150603e-11 |
| Go:0045202~synapse | 1.9075702622500652 | 2.367703511030328e-08 | 4.519232210675739e-07 | 8.662704964468304e-08 | 1.7325409173984951e-07 | 3.088308178506697e-10 | 1.3452859175467399e-10 |
| Go:0005773~vacuole | 2.1489826699910735 | 3.972453044287505e-08 | 8.996030698149582e-07 | 1.1496047025438827e-07 | 3.4488137101718053e-07 | 6.14761858975789e-10 | 2.2570753288854742e-10 |
| Go:0044456~synapse part | 2.098752999022541 | 9.238784959286761e-08 | 2.021596878787335e-06 | 1.937552197572856e-07 | 7.750206539869353e-07 | 1.3814990122642384e-09 | 5.249309998915088e-10 |
| Go:0005764~lysosome | 2.2055632970560737 | 1.0983283904586472e-07 | 2.5850706908414622e-06 | 1.9820803853765057e-07 | 9.910397996693021e-07 | 1.7665601879495592e-09 | 6.240502924891176e-10 |
| Go:0000323~lytic vacuole | 2.193241714390956 | 1.41008180132296e-07 | 3.2915711489778232e-06 | 2.1031528640858e-07 | 1.2618910548711781e-06 | 2.249361552101565e-09 | 8.011828502785253e-10 |
| Mitochondrion | 1.5099491648511256 | 3.9544751817555124e-07 | 4.728671154641262e-06 | 3.7922549811497674e-07 | 1.896126052503e-06 | 3.208338161201612e-09 | 1.938468619622939e-09 |
| Mmu04142:lysosome | 2.569262649297637 | 2.117432660320162e-07 | 4.085418514598871e-06 | 6.135593719758248e-07 | 6.135593719758248e-07 | 3.2987073620093233e-09 | 9.537986329443383e-10 |
| Lysosome | 2.296039094650206 | 1.0611784829750448e-06 | 1.15652372012498e-05 | 7.729148396862229e-07 | 4.637480077396283e-06 | 7.846854191101906e-09 | 2.600928986061933e-09 |
| Go:0031090~organelle membrane | 1.4800992756140203 | 1.3494592169260855e-06 | 1.8065630424946022e-05 | 9.894042859448504e-07 | 6.925809444391362e-06 | 1.2345514024761447e-08 | 7.667387047606795e-09 |
| Synapse | 2.0087964241512983 | 3.043611162323323e-06 | 2.725803323055942e-05 | 1.5614378080508473e-06 | 1.0930013456644794e-05 | 1.8494201550127085e-08 | 7.459842655744907e-09 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain cortex | 2.361057955999756 | 1.5772987731606894e-23 | 4.131567626707525e-22 | 7.655228987694086e-23 | 7.655228987694086e-23 | 3.2030246810435503e-25 | 1.1347473188206398e-25 |
| Hippocampus | 1.8325064476005086 | 3.737103919909007e-17 | 7.062109998442186e-16 | 6.542561330097517e-17 | 1.3085122660195034e-16 | 5.474946719746876e-19 | 2.688563971157559e-19 |
| Brain | 1.2334196829295694 | 1.437169863047729e-16 | 1.692803922994553e-15 | 1.0455112790619806e-16 | 3.136533837185942e-16 | 1.3123572540526952e-18 | 1.0339351532717475e-18 |
| Phosphoprotein | 1.2377000475360482 | 1.14832888632524e-15 | 1.049796194684422e-14 | 3.956526470303121e-15 | 3.956526470303121e-15 | 7.19368449146022e-18 | 5.62906316826098e-18 |
| Medulla oblongata | 2.0321663621678434 | 6.141975816831291e-12 | 1.3274936705442997e-10 | 6.149303288793817e-12 | 2.459732417747773e-11 | 1.0287047945455495e-13 | 4.413183647160894e-14 |
| Visual cortex | 2.6264421228047685 | 1.1481482431463519e-11 | 3.4970915052667806e-10 | 1.2959411321844527e-11 | 6.479683456461771e-11 | 2.711065193886192e-13 | 8.257394128164157e-14 |
| Membrane | 1.2250515305294996 | 6.8421268650809e-11 | 6.065925539644468e-10 | 1.1430856261540612e-10 | 2.2861712523081223e-10 | 4.156326759698775e-13 | 3.3536316463114063e-13 |
| Liver | 1.3299924573029471 | 5.706790595638722e-11 | 7.43105577072356e-10 | 2.2947754807489673e-11 | 1.376866398672405e-10 | 5.761473676115099e-13 | 4.1053513832472316e-13 |
| Kidney | 1.4347271717301202 | 8.17592660240507e-11 | 1.1369238883673916e-09 | 3.009370530548949e-11 | 2.1065604816072891e-10 | 8.814598572853822e-13 | 5.88222851922757e-13 |
| Eye | 1.536674768214871 | 1.0953704610017212e-10 | 1.6674994718357539e-09 | 3.8620662223820545e-11 | 3.089657418797742e-10 | 1.2926982540620987e-12 | 7.880240739864171e-13 |
| Colon | 1.6125587767304796 | 3.6152163485070332e-09 | 5.684867021571449e-08 | 1.170364472002916e-09 | 1.053328002598164e-08 | 4.407232684193411e-11 | 2.6008732790724088e-11 |
| Cytoplasm | 1.2682986222243504 | 1.051646239424997e-06 | 1.0287057716773518e-05 | 1.2923458269087362e-06 | 3.877032470400721e-06 | 7.049163568917332e-09 | 5.155131355319011e-09 |
| Go:0019226~transmission of nerve impulse | 2.250627040840471 | 1.0587544015949746e-06 | 1.557323857648285e-05 | 2.7049496305053822e-05 | 2.7049496305053822e-05 | 8.603645675168872e-09 | 3.208348415091659e-09 |
| Alternative splicing | 1.2051984545434413 | 1.4734405218153057e-06 | 1.3789251929186008e-05 | 1.2992404821998704e-06 | 5.196951800678917e-06 | 9.4490277439548e-09 | 7.222752945063708e-09 |
| Mammary tumor | 1.3030215479067007 | 1.2469273251713986e-06 | 1.555248329010439e-05 | 2.881661967935045e-07 | 2.8816582313684336e-06 | 1.2057164757346623e-08 | 8.970705709974038e-09 |
| Go:0044456~synapse part | 2.217335792310966 | 1.1411830405094747e-06 | 2.5001597159235445e-05 | 8.97855160897354e-06 | 8.97855160897354e-06 | 1.7266522804811748e-08 | 6.483998232394569e-09 |
| Go:0005739~mitochondrion | 1.4158501741908065 | 2.8803870986138236e-06 | 3.487081823294602e-05 | 6.261397621942777e-06 | 1.252275603880193e-05 | 2.4082373634181522e-08 | 1.6365859262142943e-08 |
| Go:0045202~synapse | 1.9290631823217206 | 3.6779701293454536e-06 | 6.719059838511399e-05 | 8.043148348568785e-06 | 2.412925096961782e-05 | 4.640296445332709e-08 | 2.0897595771899867e-08 |
| Voltage-gated channel | 2.5238903025162567 | 3.4440605303220906e-06 | 7.835237066933232e-05 | 5.905954625462506e-06 | 2.9529424326102394e-05 | 5.369065372382776e-08 | 1.688267858630203e-08 |
| Go:0048878~chemical homeostasis | 1.8663467877811148 | 1.7761401619464934e-05 | 0.00020482171595448406 | 0.00017786639741523835 | 0.00035570115837502136 | 1.1315662047042635e-07 | 5.382290570401704e-08 |
| Potassium | 2.598751455556993 | 2.3034184656656187e-05 | 0.00018199130132900976 | 1.1431583010423907e-05 | 6.858753787630789e-05 | 1.2470888329835185e-07 | 3.763798939687441e-08 |
| Synapse | 2.119534637852561 | 2.786458464099084e-05 | 0.0002504767657618423 | 1.3485781032329669e-05 | 9.439664812016524e-05 | 1.7163835572538126e-07 | 6.829649977475203e-08 |
| Corpora quadrigemina | 1.6740871503607349 | 1.7414467317955484e-05 | 0.0002947093715843785 | 4.964141399388389e-06 | 5.4604200065311304e-05 | 2.284756682090344e-07 | 1.252850205968984e-07 |
| Kinase | 1.5542273556690456 | 3.437840963305128e-05 | 0.00041978746520010546 | 1.9776306215302242e-05 | 0.00015819949929152255 | 2.8765816623075574e-07 | 1.685244985172628e-07 |
| Go:0008021~synaptic vesicle | 3.1422138233283747 | 1.2805275822702633e-05 | 0.0004595656862949049 | 4.1259130092008434e-05 | 0.00016502630675407914 | 3.1738442091972557e-07 | 7.275771218741473e-08 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0006695~cholesterol biosynthetic process | 22.353936545240895 | 0.0002500155405384952 | 0.0007693850802459323 | 0.0005900467218694638 | 0.0005900467218694638 | 4.736923981895849e-07 | 1.5785782626577753e-08 |
| Cholesterol biosynthesis | 26.687593423019436 | 5.9297967790583606e-05 | 0.002768180803269793 | 0.0005289873116460786 | 0.0005289873116460786 | 2.133575444919508e-06 | 5.8136984576270105e-08 |
| Go:0016126~sterol biosynthetic process | 17.13801801801802 | 0.0013161807166904849 | 0.004182585776890679 | 0.0016030414193964893 | 0.003203513097000621 | 2.5751614756825803e-06 | 1.2094100677597368e-07 |
| Mmu04010:mapk signaling pathway | 4.511006289308176 | 7.47244256255053e-05 | 0.003483787404567895 | 0.0002855914764230416 | 0.0002855914764230416 | 3.209346147041784e-06 | 6.732179502821933e-07 |
| Go:0006694~steroid biosynthetic process | 9.310392082223068 | 0.0011001470934980384 | 0.008101195375365133 | 0.0020694969914446526 | 0.006195651384219847 | 4.987888104690003e-06 | 4.765162066215539e-07 |
| Steroid biosynthesis | 14.010986547085203 | 0.0008090926441417512 | 0.012082844285055128 | 0.0011541821639248218 | 0.0023070321913821745 | 9.313252962902658e-06 | 5.668206630372846e-07 |
| Sterol biosynthesis | 20.015695067264573 | 0.0006458982771379018 | 0.012920911404823432 | 0.0008229640650959658 | 0.002466860943099225 | 9.95925951018639e-06 | 3.958987937742925e-07 |
| Lipid synthesis | 7.27843456991439 | 0.0008391283115307679 | 0.04236285901296766 | 0.0020227274194584144 | 0.008066394207173144 | 3.265714247571041e-05 | 4.115092399534614e-06 |
| Go:0008203~cholesterol metabolic process | 8.394131274131276 | 0.016775330396020305 | 0.0706026188302511 | 0.013453811446580866 | 0.05273892359970456 | 4.34826307942612e-05 | 4.660493247362743e-06 |
| Go:0033673~negative regulation of kinase activity | 10.081187069422365 | 0.02465642047505323 | 0.1001706099950983 | 0.015258869871866754 | 0.07400127573382331 | 6.170148484350231e-05 | 5.403769123008026e-06 |
| Go:0006469~negative regulation of protein kinase activity | 10.081187069422365 | 0.02465642047505323 | 0.1001706099950983 | 0.015258869871866754 | 0.07400127573382331 | 6.170148484350231e-05 | 5.403769123008026e-06 |
| Go:0043086~negative regulation of catalytic activity | 6.544929087503345 | 0.027569519328653036 | 0.10892204164074659 | 0.013837196328476353 | 0.08020359868078819 | 6.709480870878911e-05 | 9.412983042898556e-06 |
| Go:0008202~steroid metabolic process | 5.018230653013262 | 0.020477646123489013 | 0.10938750905250538 | 0.011922697555778683 | 0.08053233433385254 | 6.738167893617328e-05 | 1.2539448735729754e-05 |
| Go:0051348~negative regulation of transferase activity | 9.700764915859256 | 0.02971457280105494 | 0.12484881084243726 | 0.011907925371710482 | 0.09138620317310397 | 7.691128058894833e-05 | 7.031442860859898e-06 |
| Go:0016125~sterol metabolic process | 7.631028431028431 | 0.0031560664669987304 | 0.13076129773634815 | 0.01109099115481349 | 0.09550327328020602 | 8.055581863835825e-05 | 9.57891578420833e-06 |
| Go:0008299~isoprenoid biosynthetic process | 16.692874692874692 | 0.15829336475213862 | 0.3189835417259812 | 0.024211390573047886 | 0.21736828691129295 | 0.00019668454534437545 | 9.670196769954713e-06 |
| Cleavage on pair of basic residues | 4.036778837095376 | 0.009522729333161006 | 0.2598115291848391 | 0.009895895604108529 | 0.048509833594017726 | 0.00020048767035711152 | 4.690262768274462e-05 |
| Go:0045859~regulation of protein kinase activity | 4.34373728567277 | 0.06132185184237082 | 0.3590280884000929 | 0.024771383494908927 | 0.24112402978138114 | 0.00022141764002539822 | 4.7940228667717125e-05 |
| Go:0006793~phosphorus metabolic process | 2.205155733100306 | 0.0742603146692371 | 0.4345878312561391 | 0.027458633664401044 | 0.28402642547974233 | 0.000268111740062562 | 0.0001169055920847239 |
| Go:0006796~phosphate metabolic process | 2.205155733100306 | 0.0742603146692371 | 0.4345878312561391 | 0.027458633664401044 | 0.28402642547974233 | 0.000268111740062562 | 0.0001169055920847239 |
| Go:0043549~regulation of kinase activity | 4.207995495495496 | 0.04128217565165859 | 0.46362581344323583 | 0.027049612094676934 | 0.29986942774878556 | 0.0002860653592451622 | 6.387445519101104e-05 |
| Go:0051338~regulation of transferase activity | 4.05997555344289 | 0.028821987917542646 | 0.6210111645761573 | 0.033558412023238726 | 0.3799045513499022 | 0.00038345925633032464 | 8.861879700492282e-05 |
| Go:0044092~negative regulation of molecular function | 5.007862407862409 | 0.07586702569844761 | 0.6937082418686846 | 0.0349749967834615 | 0.41375589730897266 | 0.0004284948621768484 | 7.969309313774922e-05 |
| Go:0016311~dephosphorylation | 4.688211615871191 | 0.1961640714472105 | 1.0739037407353824 | 0.0504503618662806 | 0.563199867246901 | 0.000664529945310235 | 0.00013233069781904842 |
| Go:0006470~protein amino acid dephosphorylation | 5.154291133238502 | 0.14934269155283808 | 1.4620059172892996 | 0.06430011762250076 | 0.6769112969113118 | 0.000906353356058455 | 0.00016336637576297043 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mmu04010:mapk signaling pathway | 7.466493168510085 | 3.805039234383578e-05 | 0.0025016864430460473 | 0.0001197006534521261 | 0.0001197006534521261 | 2.6601701983102284e-06 | 3.4280279062665396e-07 |
| Go:0006470~protein amino acid dephosphorylation | 11.275011853959223 | 0.0027201849744982187 | 0.04935463123355577 | 0.021848295927252237 | 0.021848295927252237 | 3.292122311874331e-05 | 2.751401670065855e-06 |
| Go:0016311~dephosphorylation | 9.115967030860649 | 0.018382484583779002 | 0.1617152995956994 | 0.03556342665561585 | 0.06986209599574222 | 0.0001079262470073923 | 1.1244507691268489e-05 |
| Protein phosphatase | 10.886585365853657 | 0.01142110574145827 | 0.25791899778390137 | 0.03278217070870859 | 0.03278217070870859 | 0.00021501911186656348 | 1.8769149770053407e-05 |
| Go:0006695~cholesterol biosynthetic process | 31.934195064629847 | 0.09976650106554563 | 0.3650941636190397 | 0.053095190348796506 | 0.15097795394252778 | 0.00024388996476631592 | 6.6351488236134e-06 |
| Lipoprotein | 4.066296741065883 | 0.015552536479002343 | 0.38758912955392244 | 0.024749448092266535 | 0.048886361003661394 | 0.00032331371874071645 | 7.683403802530594e-05 |
| Go:0016126~sterol biosynthetic process | 24.482882882882883 | 0.19528619819263937 | 0.8129878360820353 | 0.0872744213265344 | 0.30599773742507475 | 0.0005442334582013565 | 1.9951003040611612e-05 |
| Go:0006694~steroid biosynthetic process | 12.931100114198706 | 0.09061454899327526 | 0.8477592066498785 | 0.07336352700338089 | 0.3168014254680629 | 0.0005676029701395276 | 4.1118737360092555e-05 |
| Go:0008202~steroid metabolic process | 6.843041799563538 | 0.32720911002369313 | 2.55275689893526 | 0.17539881592766715 | 0.6856136189542436 | 0.0017230035445646372 | 0.0002401655258709924 |
| Go:0006793~phosphorus metabolic process | 2.7564446663753825 | 0.381468999642131 | 3.079003348235887 | 0.18120365893178592 | 0.7532647197594543 | 0.00208342911179322 | 0.0007276260101464184 |
| Go:0006796~phosphate metabolic process | 2.7564446663753825 | 0.381468999642131 | 3.079003348235887 | 0.18120365893178592 | 0.7532647197594543 | 0.00208342911179322 | 0.0007276260101464184 |
| Go:0043086~negative regulation of catalytic activity | 9.09017928819909 | 0.4803873357735806 | 3.122540250696748 | 0.162591314385241 | 0.7581758395471094 | 0.0021133292330561583 | 0.0002204038628704016 |
| Go:0033673~negative regulation of kinase activity | 14.40169581346052 | 0.5374541797781792 | 3.7926444016697647 | 0.17488942584077483 | 0.8227405589761441 | 0.0025751299925202385 | 0.00016687128240538455 |
| Go:0006469~negative regulation of protein kinase activity | 14.40169581346052 | 0.5374541797781792 | 3.7926444016697647 | 0.17488942584077483 | 0.8227405589761441 | 0.0025751299925202385 | 0.00016687128240538455 |
| Go:0051348~negative regulation of transferase activity | 13.85823559408465 | 0.5648479674107629 | 4.2249325925591386 | 0.17565475756593862 | 0.8550923350505585 | 0.0028746360553073514 | 0.00019393452764059885 |
| Cholesterol biosynthesis | 36.28861788617886 | 0.0712805994124498 | 3.505737094018546 | 0.14232572598816384 | 0.36909037817885315 | 0.0029671547967564426 | 7.249602926170564e-05 |
| Go:0045859~regulation of protein kinase activity | 5.923278116826505 | 0.4965113116254988 | 4.7284679943053405 | 0.17884961399946764 | 0.8855410611972455 | 0.003225099830903244 | 0.0005197088689800512 |
| Go:0043549~regulation of kinase activity | 5.738175675675676 | 0.33345430878151894 | 5.3979976107061844 | 0.18692057940846296 | 0.9165174650111204 | 0.0036937838331197125 | 0.0006144272042907608 |
| Go:0051338~regulation of transferase activity | 5.5363303001493955 | 0.2170831859721989 | 6.257906886128928 | 0.19943716338603268 | 0.944519478730341 | 0.004300299826346266 | 0.0007413275920399324 |
| Cleavage on pair of basic residues | 5.489034638245542 | 0.14935585913111016 | 5.324104412358032 | 0.1618282860185808 | 0.5064490389107901 | 0.004545310671840904 | 0.0007926337571514524 |
| Sterol biosynthesis | 27.21646341463414 | 0.2500219067874746 | 6.136104171461576 | 0.1507998002746589 | 0.5583782664675658 | 0.005259035948120615 | 0.00017627813883497282 |
| Go:0044092~negative regulation of molecular function | 6.955364455364457 | 0.5267348965199066 | 7.9676314746784715 | 0.23308951444288506 | 0.9756541209783568 | 0.005521796416854807 | 0.0007553707001898547 |
| Go:0051726~regulation of cell cycle | 5.148269765092196 | 0.6573127483399668 | 8.400709385710181 | 0.23030732063926573 | 0.9802866542378059 | 0.005834565809861496 | 0.0010811697108827172 |
| Go:0008203~cholesterol metabolic process | 10.492664092664093 | 0.8720821109063241 | 9.033846197775441 | 0.23264205644574287 | 0.9855466709046562 | 0.006294310741647108 | 0.0005663317808466789 |
| Metal-binding | 1.7860169876866554 | 0.5545267671286324 | 8.454839324636843 | 0.17306006641647442 | 0.6802255289985548 | 0.007328750256364992 | 0.003955968757698925 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mmu04010:mapk signaling pathway | 13.324818577648767 | 2.1792023449096476e-06 | 0.00020477224914605685 | 5.381003744475699e-06 | 5.381003744475699e-06 | 2.6905087485938495e-07 | 1.963247470795819e-08 |
| Dna-binding | 4.114169599463717 | 0.007304035985288171 | 0.16856363395155727 | 0.015448421399170242 | 0.015448421399170242 | 0.00015114383505930026 | 3.593484971690957e-05 |
| Mammary gland | 3.401472350624893 | 0.012311746328884432 | 0.2925689733029202 | 0.014158410674483757 | 0.014158410674483757 | 0.0003099432011562192 | 8.911950881295169e-05 |
| Go:0045941~positive regulation of transcription | 6.459490662139219 | 0.025211041869452444 | 0.7162496074851088 | 0.18949840010699748 | 0.18949840010699748 | 0.0005135648135533074 | 7.737362654625999e-05 |
| Go:0010628~positive regulation of gene expression | 6.2874140666314124 | 0.02980398053258193 | 0.8258265321601121 | 0.11413369065380152 | 0.2152408819653453 | 0.0005924368221066855 | 9.16841178917294e-05 |
| Go:0045935~positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 6.016192283364958 | 0.039084356351408944 | 1.0404982198584212 | 0.0968845718225565 | 0.2634032733804329 | 0.0007471894651562355 | 0.0001208068041697882 |
| Go:0051173~positive regulation of nitrogen compound metabolic process | 5.833190236722679 | 0.04717131389306983 | 1.2219834654454753 | 0.08592417105647576 | 0.30188189701214263 | 0.0008782619957222638 | 0.0001464139902106176 |
| Go:0010557~positive regulation of macromolecule biosynthetic process | 5.789166159464394 | 0.04938541950890796 | 1.2709455549673954 | 0.07204468164215938 | 0.31192571767544763 | 0.000913661765926457 | 0.00015346269830973912 |
| Go:0007611~learning or memory | 19.69989126495107 | 0.0750283865189233 | 1.3493171661259074 | 0.064035967053654 | 0.32771261759873993 | 0.0009703586710634126 | 4.726690110851802e-05 |
| Go:0045944~positive regulation of transcription from rna polymerase ii promoter | 7.34618850243287 | 0.04363563899003242 | 1.4211250941877251 | 0.058013794608212765 | 0.341869829224127 | 0.0010223438942702368 | 0.00013519179342953613 |
| Go:0031328~positive regulation of cellular biosynthetic process | 5.558438522674146 | 0.06305733799846769 | 1.5677786389955095 | 0.05609846663899831 | 0.36989411939095873 | 0.001128622795015485 | 0.0001973538316068371 |
| Go:0009891~positive regulation of biosynthetic process | 5.508542306133086 | 0.06653800700571044 | 1.6420720035918324 | 0.05235063268233664 | 0.3836475978341405 | 0.0011825188489112473 | 0.00020862984391995792 |
| Go:0007610~behavior | 6.493667861409797 | 0.08350449326684217 | 2.4451406204283654 | 0.0698002343119195 | 0.514977084575567 | 0.0017675293308511146 | 0.0002642018027110229 |
| Go:0006470~protein amino acid dephosphorylation | 15.379739671760044 | 0.1156565708881453 | 2.734981857723917 | 0.07103512362154285 | 0.5553766846258383 | 0.0019797683225452763 | 0.00012414360396770582 |
| Go:0045893~positive regulation of transcription, dna-dependent | 6.321960297766749 | 0.09581987088034682 | 2.746523625285846 | 0.06558352157721026 | 0.556916208481907 | 0.0019882320438240012 | 0.0003051857866846118 |
| Go:0051254~positive regulation of rna metabolic process | 6.276695665563168 | 0.09939585452427935 | 2.8331614440639585 | 0.06257537489608178 | 0.5683093414318741 | 0.002051794424186541 | 0.0003171904969082482 |
| Go:0010604~positive regulation of macromolecule metabolic process | 4.8471691382561275 | 0.13943660890431553 | 3.1343475064991466 | 0.06432275091771489 | 0.6057561050757025 | 0.0022731716861178982 | 0.00045495101868167794 |
| Go:0006355~regulation of transcription, dna-dependent | 2.9919630078168007 | 0.3015108951157329 | 4.59312340480097 | 0.0875486580729542 | 0.7469853237243498 | 0.003354526689407653 | 0.001086789919497096 |
| Go:0016311~dephosphorylation | 12.434683138869824 | 0.37125290797466204 | 4.944454122041897 | 0.08847263629435331 | 0.7728486257152731 | 0.0036172527692237798 | 0.0002811884485631821 |
| Go:0051252~regulation of rna metabolic process | 2.9457162677766213 | 0.3330910632874784 | 5.094821260631144 | 0.08598340372465452 | 0.783120059322798 | 0.003729973362283006 | 0.0012268279488711724 |
| M_dsppathway:regulation of map kinase pathways through dual specificity phosphatases | 27.023076923076925 | 0.05450307328279891 | 3.7473745866074437 | 0.15647216986684642 | 0.15647216986684642 | 0.0041417014453541175 | 0.0001228973626761843 |
| Go:0006357~regulation of transcription from rna polymerase ii promoter | 4.269375785504818 | 0.5419701981724894 | 13.698777642605265 | 0.21276543852812735 | 0.9865142612147314 | 0.01047318717923079 | 0.0023633272600756604 |
| Dna binding | 8.141358869129046 | 0.24933197508913674 | 12.428044836513452 | 0.4579262125491349 | 0.7061560089586744 | 0.01181994148713435 | 0.0014048542719912959 |
| Nucleus | 1.9305795847750864 | 0.7611668132686655 | 14.556919979885452 | 0.3836503800698846 | 0.7658568819280503 | 0.013996491784740073 | 0.00699497894641124 |
| Lipid synthesis | 15.912655971479502 | 0.1632860508770988 | 14.918762049339485 | 0.3111603982782468 | 0.7748497529417258 | 0.0143713373403803 | 0.0008735056452865466 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mmu04010:mapk signaling pathway | 5.249170954831332 | 0.010415187165473072 | 0.5074982959160868 | 0.028442428843221235 | 0.028442428843221235 | 0.0005151307005216033 | 9.431811747411339e-05 |
| Go:0043086~negative regulation of catalytic activity | 8.96897689768977 | 0.14971013217209095 | 0.7974199040983954 | 0.34560693440553514 | 0.34560693440553514 | 0.0005201678440993293 | 5.460388513283175e-05 |
| Cell adhesion | 4.121421975992613 | 0.032450691125769926 | 0.840331268905159 | 0.10837663952760102 | 0.10837663952760102 | 0.0006949794637268842 | 0.00016169717134907242 |
| Calcium | 2.9994480044159646 | 0.0462452265066331 | 0.8740007299369856 | 0.05791890538444722 | 0.1124832111679619 | 0.000722937589865608 | 0.00023207449508941733 |
| Cleavage on pair of basic residues | 5.264337313872917 | 0.029120359332572177 | 0.9649230185663549 | 0.04298284411258946 | 0.12348536954800227 | 0.0007984803348300084 | 0.0001448560385873401 |
| Go:0007156~homophilic cell adhesion | 7.742450142450143 | 0.15079999536580624 | 1.5487091410506193 | 0.33856703483614115 | 0.5625064325945455 | 0.0010138333017579103 | 0.00012382607814243922 |
| Go:0007610~behavior | 3.7278463648834017 | 0.10556681959754244 | 2.004507544987899 | 0.300571304567404 | 0.657839132483843 | 0.001315053919316965 | 0.0003380188567005469 |
| Go:0010648~negative regulation of cell communication | 5.6819593787335725 | 0.2625824870875072 | 2.0980409677891054 | 0.24479266814833212 | 0.6747144356247998 | 0.001377028721398745 | 0.00023073174394341368 |
| Go:0044092~negative regulation of molecular function | 6.8626262626262635 | 0.21109510169839385 | 2.634633801634545 | 0.24635175468958426 | 0.7568672503421134 | 0.0017336460363844924 | 0.00023947586640241274 |
| Go:0045944~positive regulation of transcription from rna polymerase ii promoter | 3.7955307262569833 | 0.1788712076041712 | 3.5857335343260743 | 0.2755521661874397 | 0.8554418076102673 | 0.002370282307665031 | 0.0005970196196215312 |
| Dna-binding | 2.119420702754036 | 0.2473554505661485 | 3.6581893000470767 | 0.11895637889946964 | 0.3974547811599719 | 0.003065549544096704 | 0.0013919822952886927 |
| Go:0045941~positive regulation of transcription | 3.1784795321637422 | 0.31819481105144665 | 5.773766695662652 | 0.36236419882410453 | 0.9571442743579905 | 0.0038574684147661296 | 0.0011599670467495622 |
| Go:0006469~negative regulation of protein kinase activity | 11.841394335511984 | 0.8057065189515819 | 6.716681914332501 | 0.36892177206696297 | 0.9748426102033411 | 0.004508338813240931 | 0.00035456601711005925 |
| Go:0033673~negative regulation of kinase activity | 11.841394335511984 | 0.8057065189515819 | 6.716681914332501 | 0.36892177206696297 | 0.9748426102033411 | 0.004508338813240931 | 0.00035456601711005925 |
| Go:0016337~cell-cell adhesion | 4.478154425612053 | 0.6132063949953519 | 6.7204119212304185 | 0.335960404059659 | 0.9748958344787132 | 0.004510925735903017 | 0.000958998501704655 |
| Go:0010628~positive regulation of gene expression | 3.093806921675774 | 0.37436984018562547 | 6.848600163105367 | 0.3132299860635953 | 0.9766593339244222 | 0.004599888710703259 | 0.001420190214927792 |
| Go:0051348~negative regulation of transferase activity | 11.39454926624738 | 0.828787917738742 | 7.45745462866636 | 0.31145269723949875 | 0.9835080657347991 | 0.005024004560128272 | 0.0004113028611969794 |
| Go:0009968~negative regulation of signal transduction | 5.297465886939571 | 0.4646842961858386 | 7.820785493905513 | 0.301930486599841 | 0.9866098926396255 | 0.005278338273760068 | 0.0009463679800958915 |
| Go:0005624~membrane fraction | 3.2449826989619375 | 0.256424394314931 | 6.4960714079130515 | 0.5407442864873042 | 0.5407442864873042 | 0.0057053396875964765 | 0.0016820203460216985 |
| Go:0045893~positive regulation of transcription, dna-dependent | 3.266346153846154 | 0.4318630611792479 | 8.652806890628762 | 0.3083873132846764 | 0.9917164143710135 | 0.005864307192423351 | 0.001711844634369898 |
| Dna binding | 4.2492180062559495 | 0.23978218408649332 | 7.040522436532615 | 0.18002383751758277 | 0.6293140409690858 | 0.005996494563846733 | 0.001342971361047423 |
| Go:0045935~positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 2.9603485838779955 | 0.47800498945497505 | 8.994524861147601 | 0.29992685405504704 | 0.9932078406761365 | 0.0061064162452595825 | 0.001968052505120336 |
| Go:0051254~positive regulation of rna metabolic process | 3.242959427207637 | 0.44783371377280734 | 9.009054319501352 | 0.2834898590813164 | 0.9932650384360805 | 0.006116729274721941 | 0.0017980967408843403 |
| Go:0005576~extracellular region | 1.9701680672268906 | 0.41670833569657706 | 7.105501356274013 | 0.34750539914645473 | 0.5742507958569727 | 0.00625904205283289 | 0.003058200149443394 |
| Go:0005626~insoluble fraction | 3.1343582887700534 | 0.3131828859410526 | 7.907548415201782 | 0.27248604545427313 | 0.6149439211083749 | 0.006992834781243278 | 0.0021323099341743782 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mmu04010:mapk signaling pathway | 11.659216255442669 | 6.216241517553733e-05 | 0.00502284624493976 | 0.00012733503075224029 | 0.00012733503075224029 | 6.702247989556685e-06 | 5.600390083809429e-07 |
| Mammary gland | 3.684928379843634 | 0.0023212851851717176 | 0.05873131197497994 | 0.0027591262492177027 | 0.0027591262492177027 | 6.279211154067217e-05 | 1.6719166306632962e-05 |
| Dna-binding | 4.3713051994302 | 0.003821052207465314 | 0.09105788315078023 | 0.007508333478228546 | 0.007508333478228546 | 8.373719281953885e-05 | 1.8766348798421094e-05 |
| Go:0045944~positive regulation of transcription from rna polymerase ii promoter | 8.856238361266294 | 0.0033035392343636616 | 0.12635742138054074 | 0.034411164228033475 | 0.034411164228033475 | 9.118639233356118e-05 | 1.0027246598202198e-05 |
| Go:0045893~positive regulation of transcription, dna-dependent | 7.621474358974359 | 0.00870837827310389 | 0.28712144085352165 | 0.03903446447742265 | 0.07654523953780612 | 0.00020735732034653156 | 2.6504248098061906e-05 |
| Go:0051254~positive regulation of rna metabolic process | 7.566905330151154 | 0.009117740439435473 | 0.29851258056081686 | 0.02722180260333773 | 0.07946250027720136 | 0.00021559535158276798 | 2.7755861579401005e-05 |
| Go:0045941~positive regulation of transcription | 6.674807017543859 | 0.02017002030441839 | 0.5851805894688478 | 0.03982129935765033 | 0.15002085183024438 | 0.0004232006892832671 | 6.174419215869818e-05 |
| Go:0010628~positive regulation of gene expression | 6.496994535519126 | 0.02387570805137862 | 0.6753200368438406 | 0.036837593024725224 | 0.17110863218343886 | 0.0004885945895548344 | 7.322565938310842e-05 |
| Go:0045935~positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 6.216732026143791 | 0.031384060656955426 | 0.8521991670405349 | 0.03873491351694247 | 0.2110326826000859 | 0.0006170761494442857 | 9.662288651110729e-05 |
| Go:0051173~positive regulation of nitrogen compound metabolic process | 6.027629911280101 | 0.03794787562423285 | 1.0019995497935197 | 0.0390604268574134 | 0.24338917384417746 | 0.0007260546746798695 | 0.0001172253905108612 |
| Go:0010557~positive regulation of macromolecule biosynthetic process | 5.982138364779875 | 0.0397482055946099 | 1.0424517701552283 | 0.03562799844353692 | 0.2519050126463158 | 0.000755509564625806 | 0.00012290075942287645 |
| Transcription regulation | 3.6089100905562743 | 0.04999811984422908 | 1.0081682307231499 | 0.04104831774877293 | 0.08041167110754166 | 0.0009310016563187147 | 0.0002513964106946701 |
| Go:0031328~positive regulation of cellular biosynthetic process | 5.743719806763285 | 0.05089459220797099 | 1.2880141692551472 | 0.03910768270716414 | 0.30165104212364213 | 0.0009345535572885075 | 0.00015827660642494897 |
| Go:0009891~positive regulation of biosynthetic process | 5.692160383004189 | 0.0537401753452349 | 1.3495564451797115 | 0.03693160322630529 | 0.3136094169191296 | 0.0009794899086585794 | 0.0001673741476207946 |
| Go:0045449~regulation of transcription | 2.6439754527765307 | 0.13781537572956482 | 1.675590019589701 | 0.04165233148404046 | 0.3737409758531798 | 0.001217984780967529 | 0.00044925011742868797 |
| Go:0007610~behavior | 6.710123456790124 | 0.0695744459286668 | 2.0725537440704045 | 0.047186356356249126 | 0.4401191569818804 | 0.0015093579442564554 | 0.00021850101145495394 |
| Go:0006357~regulation of transcription from rna polymerase ii promoter | 5.146969696969697 | 0.0974920046393013 | 2.259071519879685 | 0.04751350003973498 | 0.46891409415510377 | 0.0016466414666134336 | 0.00031079329887156803 |
| Go:0010604~positive regulation of macromolecule metabolic process | 5.0087414428646655 | 0.1140214516890965 | 2.5914607688125813 | 0.05061524430360165 | 0.5167280869624312 | 0.0018918951288799696 | 0.0003667888998586427 |
| Transcription | 3.1539711701526287 | 0.1406778937942953 | 2.593314299775562 | 0.06989587530497199 | 0.19537279740363567 | 0.0024123767665082587 | 0.0007429172784583217 |
| Activated spleen | 5.583251369959252 | 0.08119931055829022 | 3.2294975222936784 | 0.07429049990196723 | 0.14306192142825036 | 0.0035027060875291115 | 0.0006090666579117624 |
| Cleavage on pair of basic residues | 9.377100840336134 | 0.1550817354584857 | 8.35087168921974 | 0.16502819999942053 | 0.5139429662752585 | 0.007983839701961622 | 0.0008257147169687179 |
| Go:0006350~transcription | 2.5560571858540255 | 0.6854858026983313 | 12.116413403316384 | 0.21216730670964679 | 0.9720417498436356 | 0.009271963789641879 | 0.0034990943403532646 |
| Go:0006355~regulation of transcription, dna-dependent | 2.7825255972696246 | 0.6700633756514216 | 12.623302972534889 | 0.20830398775130632 | 0.9761803531332504 | 0.009685201046255414 | 0.003354526689407653 |
| Go:0005576~extracellular region | 3.2562499999999996 | 0.4048995589222044 | 9.229613476819598 | 0.4090630001122991 | 0.4090630001122991 | 0.009876299546713114 | 0.0029446620842029005 |
| Dna binding | 8.650193798449612 | 0.20342100621824188 | 10.303556352805076 | 0.16466292400142046 | 0.5932675817598019 | 0.009945763366172415 | 0.0011142267213817584 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0045449~regulation of transcription | 4.27103726986978 | 0.07564266080039705 | 1.1954869408038582 | 0.12637687931572616 | 0.12637687931572616 | 0.0010308134483903472 | 0.00023832478209376813 |
| Go:0006355~regulation of transcription, dna-dependent | 5.565051194539249 | 0.07076344554761216 | 1.454364432724986 | 0.0789943135370651 | 0.15174852550293805 | 0.001255532815107341 | 0.0002223750866194289 |
| Go:0051252~regulation of rna metabolic process | 5.479032258064516 | 0.07696254677197967 | 1.5619255437058555 | 0.057244904497237736 | 0.1620913665741175 | 0.0013490602963883184 | 0.00024265378992906873 |
| Go:0045944~positive regulation of transcription from rna polymerase ii promoter | 15.182122905027933 | 0.02854266232752556 | 1.5671774201537536 | 0.04339181583807161 | 0.1625934212055724 | 0.0013536293460692413 | 8.774743785256407e-05 |
| Go:0045893~positive regulation of transcription, dna-dependent | 13.065384615384616 | 0.05050986986907158 | 2.4049476970762984 | 0.05322432471563576 | 0.23926139265928992 | 0.002085345280276903 | 0.00015704870774411726 |
| Go:0051254~positive regulation of rna metabolic process | 12.971837708830549 | 0.0518945261694187 | 2.4544364744354064 | 0.04546142067593195 | 0.2435836369579899 | 0.0021287486248357176 | 0.00016147036710836024 |
| Go:0045941~positive regulation of transcription | 11.442526315789474 | 0.08277839768326045 | 3.49504577649834 | 0.055493244320997115 | 0.3294433401179 | 0.0030460922556160824 | 0.00026180262012144795 |
| Go:0010628~positive regulation of gene expression | 11.137704918032789 | 0.09139377513498159 | 3.769127586743737 | 0.052520162782102475 | 0.35052892483401077 | 0.0032892120695528163 | 0.0002903925995082686 |
| Go:0045935~positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 10.657254901960785 | 0.10728548698167228 | 4.261610002130045 | 0.052908414875001974 | 0.3869049617132785 | 0.003727655718261091 | 0.00034384524587249073 |
| Go:0051173~positive regulation of nitrogen compound metabolic process | 10.333079847908746 | 0.11990159453744853 | 4.642669579338799 | 0.052002767753143164 | 0.41376641699810546 | 0.004068319910949085 | 0.00038696012183187054 |
| Go:0010557~positive regulation of macromolecule biosynthetic process | 10.255094339622643 | 0.12319679922743842 | 4.740971828381523 | 0.04839204324800883 | 0.42051965304017247 | 0.004156403264999528 | 0.00039832280681932034 |
| Go:0031328~positive regulation of cellular biosynthetic process | 9.846376811594203 | 0.14233843667830903 | 5.30350409337631 | 0.049733860251101825 | 0.45782054697925256 | 0.004662062342883978 | 0.0004651817865152916 |
| Go:0009891~positive regulation of biosynthetic process | 9.757989228007181 | 0.14692988587445466 | 5.436536821049865 | 0.04715583017486302 | 0.4663156679285946 | 0.004782046632294679 | 0.00048144024549340265 |
| Dna-binding | 5.78023828023828 | 0.1715686677437609 | 4.97315315983159 | 0.21329585109334315 | 0.21329585109334315 | 0.005437504415272343 | 0.0009222280525391487 |
| Go:0006357~regulation of transcription from rna polymerase ii promoter | 8.823376623376625 | 0.2078044728579339 | 7.152554442221581 | 0.057810054157265145 | 0.5655512843414403 | 0.006343742366419918 | 0.000705651032130852 |
| Go:0010604~positive regulation of macromolecule metabolic process | 8.586413902053712 | 0.22757670945732078 | 7.6972227202827925 | 0.05822109804425657 | 0.5933372856640751 | 0.006844947941214917 | 0.000782186585216443 |
| Leucine zipper | 202.88636363636365 | 0.04919991760232589 | 8.047772737430925 | 0.17904981457329794 | 0.3260407930478635 | 0.008927765382415485 | 4.1217661014737814e-05 |
| Mmu04010:mapk signaling pathway | 12.991698113207546 | 0.09600430868606569 | 7.2360580827646075 | 0.1242135604706266 | 0.1242135604706266 | 0.01198514527977642 | 0.0009088721743288938 |
| Mammary gland | 4.251840438281116 | 0.4017989776631883 | 11.93291015477529 | 0.3003673274109141 | 0.3003673274109141 | 0.016105256994290388 | 0.0036897833234074175 |
| Liver | 3.141185770750988 | 0.5470697596046764 | 13.4842081157729 | 0.1841938887242528 | 0.33446038880514306 | 0.018336936380010414 | 0.005681762565345673 |
| Activated spleen | 8.374877054938878 | 0.4778580651097105 | 27.38685519452353 | 0.25908159454066115 | 0.5932653702364873 | 0.0400659079369518 | 0.004664023476971395 |
| Nucleus | 2.557391138273491 | 0.9627003743454287 | 33.28974326959514 | 0.4698568488280287 | 0.8510023339217561 | 0.04234600612956757 | 0.01599217656478589 |
| Go:0031981~nuclear lumen | 7.0804077010192525 | 0.6514265882561032 | 27.761751491285548 | 0.5480192171992959 | 0.5480192171992959 | 0.04315851075549517 | 0.005970211695090314 |
| Transcription regulation | 4.199459014465483 | 0.900607182605191 | 37.491526737044175 | 0.42445985829902333 | 0.890275786242599 | 0.048982096949368004 | 0.011253239643200201 |
| Go:0070013~intracellular organelle lumen | 5.518093556928508 | 0.8813240592004504 | 40.57745839139344 | 0.47033313390294307 | 0.7194530109589223 | 0.06817652954652204 | 0.012036961678703497 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.6165878171899404 | 2.4046815042557e-15 | 2.5342047123153538e-14 | 5.171464225529336e-15 | 5.171464225529336e-15 | 1.929650830421394e-17 | 1.178765443262598e-17 |
| Coiled coil | 2.4543604970856703 | 5.662137425588298e-13 | 9.181544413650045e-12 | 9.372502773885572e-13 | 1.8745005547771143e-12 | 7.00132499053255e-15 | 2.7354112297846715e-15 |
| Alternative splicing | 1.7187515041723007 | 9.602985073797754e-12 | 1.081912337497215e-10 | 7.35911331872785e-12 | 2.2077450978486013e-11 | 8.241524058373121e-14 | 4.7043631143911305e-14 |
| Brain | 1.431044707578841 | 2.9093643938438163e-08 | 3.595413522461399e-07 | 4.3413803774150495e-08 | 4.3413803774150495e-08 | 3.0359301076844195e-10 | 2.0930682271626894e-10 |
| Transcription regulation | 2.070321530940966 | 3.194888713586863e-06 | 4.3942673766395046e-05 | 2.241805481251191e-06 | 8.967191770903327e-06 | 3.345982030217804e-08 | 1.5661244048986607e-08 |
| Brain cortex | 2.960953800298063 | 2.5604302137383073e-06 | 6.754206315351396e-05 | 4.077768729349174e-06 | 8.155520830444019e-06 | 5.7031845126678824e-08 | 1.8420384634197445e-08 |
| Nucleus | 1.5759833345102747 | 8.091106681873406e-06 | 8.379310403316964e-05 | 3.419869581633428e-06 | 1.7099230953276923e-05 | 6.38036440057429e-08 | 3.9662447349088835e-08 |
| Cerebellum | 2.0173289108508765 | 1.83530403913279e-05 | 0.0003201446145073561 | 1.288551693157114e-05 | 3.865605268726924e-05 | 2.703272351067614e-07 | 1.3203746474821228e-07 |
| Go:0045449~regulation of transcription | 1.7306021145446382 | 0.00011100569117972192 | 0.0010087134451386426 | 0.001020374462112339 | 0.001020374462112339 | 5.977137549034808e-07 | 3.3639949725240576e-07 |
| Transcription | 1.837607852637946 | 0.00019395007859357438 | 0.0023664695966263416 | 8.048392927140213e-05 | 0.0004828064211115368 | 1.801949905016593e-06 | 9.508274368829265e-07 |
| Eye | 1.9452494712931778 | 0.00020478247531763305 | 0.0035041876312869036 | 0.00010577694097735346 | 0.00042304063607601083 | 2.95894767129372e-06 | 1.473405001705215e-06 |
| Go:0006350~transcription | 1.7845926533962653 | 0.0006517081149921555 | 0.006128721570164419 | 0.003096643101194063 | 0.006183697003891986 | 3.631665093567858e-06 | 1.975514924540353e-06 |
| Repressor | 3.069741451567839 | 0.0002558700634609501 | 0.0053684117380603524 | 0.00015649385216776057 | 0.001094942802452481 | 4.087837503985908e-06 | 1.254424721970659e-06 |
| Thymus | 1.597523896924812 | 0.0032403949787678243 | 0.04563209646710753 | 0.0011016387420891283 | 0.0054960709934772645 | 3.8539326993811604e-05 | 2.3349773632710942e-05 |
| Go:0043228~non-membrane-bounded organelle | 1.6654974914531813 | 0.01610957425824533 | 0.10184612456594078 | 0.01857265989121515 | 0.01857265989121515 | 7.909939232918114e-05 | 4.613741314792861e-05 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.6654974914531813 | 0.01610957425824533 | 0.10184612456594078 | 0.01857265989121515 | 0.01857265989121515 | 7.909939232918114e-05 | 4.613741314792861e-05 |
| Serine/threonine-protein kinase | 2.604750233426704 | 0.020080847499186816 | 0.360506179050879 | 0.009170217281519344 | 0.07104983047591173 | 0.000274962864064309 | 9.943235122102914e-05 |
| Potassium transport | 4.673944344092777 | 0.011502609733598734 | 0.3803159731451511 | 0.008602532022096931 | 0.07481145190936678 | 0.0002900986836391064 | 5.6710531454037934e-05 |
| Go:0007010~cytoskeleton organization | 2.728209704406024 | 0.03763687064712584 | 0.5658703850195734 | 0.17423299629919164 | 0.4369167928378621 | 0.000336201006484472 | 0.00011624604957643187 |
| Go:0000932~cytoplasmic mrna processing body | 14.01793721973094 | 0.015018329469059344 | 0.48160254727735996 | 0.04343819680373362 | 0.08498951666590726 | 0.00037469668904137213 | 2.149443743054513e-05 |
| Go:0006813~potassium ion transport | 3.7058181818181817 | 0.06401004684066125 | 0.6740608856122488 | 0.1572853508351726 | 0.4956614403006101 | 0.00040068521418045534 | 0.00010022306273667154 |
| Go:0060341~regulation of cellular localization | 3.682800677583286 | 0.16098811421397508 | 0.7106900004431038 | 0.1344303623850881 | 0.5141412042337365 | 0.00042253204365601347 | 0.0001063764058742498 |
| Dna-binding | 1.6741642525956253 | 0.0666839517200778 | 0.7669608483418022 | 0.015588652607561926 | 0.14539365660052794 | 0.0005860757022441138 | 0.0003382339201166017 |
| Potassium | 4.238237667948535 | 0.07624801299860673 | 0.7820154764620391 | 0.014459036578999607 | 0.1480355329446309 | 0.0005976215707984866 | 0.00012958581170259656 |
| Go:0010605~negative regulation of macromolecule metabolic process | 2.2459504132231403 | 0.08097051208738826 | 1.0169128502514169 | 0.1583655971930169 | 0.6445808066674832 | 0.0006054708204684056 | 0.0002558371777385021 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0005576~extracellular region | 4.253061224489795 | 0.025632396211085595 | 0.6181151684966579 | 0.03264077614327099 | 0.03264077614327099 | 0.0006379765933121881 | 0.0001475267868661014 |
| Signal | 2.8750402576489535 | 0.09782529925310424 | 1.5663549408602595 | 0.11201622971508263 | 0.11201622971508263 | 0.0014839205602535867 | 0.0005045153174418832 |
| Mmu04512:ecm-receptor interaction | 34.566265060240966 | 0.012482400003863914 | 1.0952992529330796 | 0.01596513552408907 | 0.01596513552408907 | 0.002009721696972934 | 5.657929836167976e-05 |
| Secreted | 3.8266380894060017 | 0.2727933893111095 | 6.352626819086737 | 0.21881924848146717 | 0.38975663345694034 | 0.006154699758826308 | 0.001560274864108059 |
| Mmu04510:focal adhesion | 14.48989898989899 | 0.07985477881906844 | 5.90684964471051 | 0.04351071818230334 | 0.08512825376786681 | 0.011059809537468532 | 0.0007494827117093321 |
| Hippocampus | 4.082353908112107 | 0.5899119673691935 | 21.76702036719339 | 0.6287211768046077 | 0.6287211768046077 | 0.027146989438353007 | 0.006392312108142851 |
| Go:0044421~extracellular region part | 4.615725359911407 | 0.9561806926703248 | 34.35023865307216 | 0.6757361776187711 | 0.8948529734947148 | 0.042390592812504964 | 0.008846097672439062 |
| Leucine-rich repeat | 8.468300395256916 | 0.6473138433573875 | 38.43188390119732 | 0.7037686357502015 | 0.9740048028231627 | 0.044597960695209556 | 0.0050956820137523055 |
| Calcium binding | 33.03237742830712 | 0.7415109783862358 | 46.03693902976643 | 0.6866688457823207 | 0.9903613920868714 | 0.05637339095701349 | 0.0016565945352747537 |
| Mouse | 26.176548089591567 | 0.3063998560482285 | 47.966001765050635 | 0.7324289225725953 | 0.9284057185243377 | 0.07062483096282378 | 0.0026286228885762375 |
| Mmu04350:tgf-beta signaling pathway | 21.984674329501914 | 0.6647755602190282 | 34.18709235292052 | 0.18435892400637754 | 0.45737816378867957 | 0.07357096369052281 | 0.0032767667961428088 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0007155~cell adhesion | 4.6304917689000735 | 0.0011300343864375506 | 0.023779111176791545 | 0.008658112028138931 | 0.008658112028138931 | 1.63761582642159e-05 | 3.4262770294022086e-06 |
| Go:0022610~biological adhesion | 4.622252459702742 | 0.001152316164182099 | 0.024204889340195823 | 0.004415987430510193 | 0.008812473916034014 | 1.6669415503365322e-05 | 3.4938744749762064e-06 |
| Signal | 2.133094384707288 | 0.0016715252973718009 | 0.021855243009960557 | 0.0030856307897196844 | 0.0030856307897196844 | 1.7863430990386373e-05 | 8.200573504903946e-06 |
| Hydroxylation | 17.997983870967744 | 0.0004069307861700455 | 0.023890936349146852 | 0.0016877187462837062 | 0.0033725890980007556 | 1.952749197219432e-05 | 9.975818675343632e-07 |
| Collagen | 13.71274961597542 | 0.00101443049583394 | 0.0891824941482322 | 0.004196150588845926 | 0.012535702611720767 | 7.29160548129713e-05 | 4.975210068109803e-06 |
| Extracellular matrix | 7.210459891968298 | 0.006248666699444905 | 0.14152630898351948 | 0.004993555117023929 | 0.01982510435909768 | 0.0001157404593642373 | 1.5363292102040657e-05 |
| Cell adhesion | 5.052065647990945 | 0.005793887330948433 | 0.18265885581550956 | 0.005156526893178803 | 0.025518104347080017 | 0.00014940697810164472 | 2.848359929101382e-05 |
| Go:0005578~proteinaceous extracellular matrix | 5.412987012987013 | 0.006822508983604814 | 0.2570197887965531 | 0.029374817445511514 | 0.029374817445511514 | 0.00021920315529955903 | 3.8896338390405576e-05 |
| Go:0031012~extracellular matrix | 5.2027739251040215 | 0.009272896680431408 | 0.3359824674999623 | 0.019306290330550757 | 0.038239847814774075 | 0.0002866514183286009 | 5.293131289970212e-05 |
| Mmu04512:ecm-receptor interaction | 13.29471733086191 | 0.006431321902776177 | 0.37532714543203616 | 0.016613555877275976 | 0.016613555877275976 | 0.00040852893913966866 | 2.9063054931339973e-05 |
| Secreted | 2.4335302135393 | 0.06674651027102874 | 1.0370008203115177 | 0.024264877507983118 | 0.137038086672563 | 0.0008515720143735352 | 0.00033856238984065145 |
| Go:0044421~extracellular region part | 3.0002214839424144 | 0.10469342570586349 | 1.14100607258677 | 0.04334875457692611 | 0.12449037743496205 | 0.0009770895161315316 | 0.0003141241704250014 |
| Mmu04510:focal adhesion | 6.687645687645688 | 0.022190983274928855 | 1.2866492776169691 | 0.028435393841652168 | 0.056062216060374315 | 0.0014062060193842125 | 0.00020214990224316168 |
| Go:0005576~extracellular region | 2.1265306122448977 | 0.10912590024157054 | 1.678783545458351 | 0.04785303088442372 | 0.1781057214759355 | 0.0014411921537928086 | 0.0006563309050967997 |
| Calcium | 2.888869276142565 | 0.255382369099979 | 5.1990153907957914 | 0.10222808851447585 | 0.52993047675847 | 0.004353931456848434 | 0.001444467709009293 |
| Go:0044459~plasma membrane part | 1.968961595660922 | 0.42594900817195613 | 7.307542309588333 | 0.16123604589852036 | 0.5848559747898431 | 0.0064433415601789735 | 0.003148652170946925 |
| Go:0005886~plasma membrane | 1.5981909350113066 | 0.657949733974066 | 11.183654848796188 | 0.20467319211109536 | 0.746910732913695 | 0.010052172659445177 | 0.00607690126177797 |
| Glycoprotein | 1.5464934289127836 | 0.7981184812316745 | 14.29175935128012 | 0.2385740497658646 | 0.8870148434453898 | 0.01252494122388111 | 0.007812821451017702 |
| Go:0007281~germ cell development | 7.913803145020384 | 0.9920527438405123 | 17.96308806125554 | 0.9104793290776945 | 0.9992825857724984 | 0.013541855400791858 | 0.001626597723326638 |
| Go:0044420~extracellular matrix part | 7.7664596273291915 | 0.2636179016409995 | 15.491082805137356 | 0.24313163123973525 | 0.8577204482488883 | 0.01423565314410825 | 0.0017371606344228788 |
| Go:0006935~chemotaxis | 7.332973556395034 | 0.9417190970839988 | 21.570621145367685 | 0.891504887524833 | 0.9998614390993795 | 0.016591887503670757 | 0.00215107765859651 |
| Go:0042330~taxis | 7.332973556395034 | 0.9417190970839988 | 21.570621145367685 | 0.891504887524833 | 0.9998614390993795 | 0.016591887503670757 | 0.00215107765859651 |
| Spleen | 2.3289428815004265 | 0.6446212256675552 | 17.7068312372165 | 0.7692676118822712 | 0.7692676118822712 | 0.018164214509677243 | 0.0074153272549622815 |
| Go:0030030~cell projection organization | 3.7584362898764523 | 0.9670031001664183 | 25.873876827749577 | 0.8880288849654745 | 0.9999823992969753 | 0.02040582074200506 | 0.005155364751579696 |
| Go:0042612~mhc class i protein complex | 12.759183673469387 | 0.6872501289208389 | 23.36845235527213 | 0.31985247117828075 | 0.9542042689878685 | 0.022418160798052256 | 0.0016497052688759538 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mammary tumor | 1.5599541535344534 | 2.962370571069073e-08 | 4.160931821317604e-07 | 5.854752604950164e-08 | 5.854752604950164e-08 | 3.403926166555154e-10 | 2.1312015740983578e-10 |
| Phosphoprotein | 1.2469636665055663 | 5.6416137355075335e-06 | 5.030773170267011e-05 | 1.4056572331333506e-05 | 1.4056572331333506e-05 | 3.6228533201066755e-08 | 2.7655046867532997e-08 |
| Embryo | 1.5271916676275028 | 2.536290048482659e-05 | 0.0003528930978657918 | 2.4827129176063778e-05 | 4.96536419658522e-05 | 2.8869109061111374e-07 | 1.8246920734632763e-07 |
| Coiled coil | 1.5145472862426153 | 0.00017478882568000564 | 0.0019138150259556674 | 0.00026733985021587525 | 0.0005346082298361976 | 1.378223750715351e-06 | 8.56882490863415e-07 |
| Liver | 1.3515646258503402 | 0.00020890807732110162 | 0.0024792239882054368 | 0.00011627667493152583 | 0.0003487894655712642 | 2.0281974011259903e-06 | 1.5030916880267353e-06 |
| Bone marrow | 1.598829084222117 | 0.00022246157949989964 | 0.0033188467495559593 | 0.00011674197152222732 | 0.000466886120325416 | 2.7150843698841516e-06 | 1.600619781991398e-06 |
| Go:0005794~golgi apparatus | 1.8779936461388074 | 0.00033709803031378716 | 0.005300460887869285 | 0.0014962224965829884 | 0.0014962224965829884 | 3.8100257068203898e-06 | 1.9156507822913833e-06 |
| Go:0043228~non-membrane-bounded organelle | 1.4592886920270973 | 0.0010583155903444164 | 0.006337822256330394 | 0.0008947991978854875 | 0.0017887977301663804 | 4.555713641295025e-06 | 3.0081659348632982e-06 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.4592886920270973 | 0.0010583155903444164 | 0.006337822256330394 | 0.0008947991978854875 | 0.0017887977301663804 | 4.555713641295025e-06 | 3.0081659348632982e-06 |
| Visual cortex | 2.718664477285167 | 0.00022251941721995738 | 0.005817254505335878 | 0.0001636978984108639 | 0.0008182215658975034 | 4.759038047119881e-06 | 1.6010359725771003e-06 |
| Cytoplasm | 1.342677579149467 | 0.0007015483476900952 | 0.006697828886803592 | 0.0006236479773304993 | 0.0018697773641522808 | 4.8235130571715385e-06 | 3.4401634350601048e-06 |
| Acetylation | 1.390641487251084 | 0.0013623199776553463 | 0.013414099089470444 | 0.0009366445669947154 | 0.0037413177358246452 | 9.660607899125942e-06 | 6.682569726238242e-06 |
| Kidney | 1.4292407537727736 | 0.0011550837223046617 | 0.014923271075939493 | 0.0003499348103087607 | 0.0020977729030764447 | 1.2209090065533015e-05 | 8.314723314991709e-06 |
| Golgi apparatus | 1.8385942076147088 | 0.005012978980333815 | 0.06703108622759801 | 0.0037401245263462 | 0.018561259527838603 | 4.8286672830732675e-05 | 2.4634922584523394e-05 |
| Salivary gland | 1.8430426716141002 | 0.008603386558311055 | 0.14620656673336452 | 0.0029367433156282097 | 0.020376973397311815 | 0.00011968730232588509 | 6.216072504173824e-05 |
| Go:0005739~mitochondrion | 1.461453605648008 | 0.024295613581143627 | 0.2996297588236829 | 0.027860570646440097 | 0.08127470344153997 | 0.00021567169350106817 | 0.00013973808353611597 |
| Eye | 1.4397492685489641 | 0.02432103481123149 | 0.32127989603526474 | 0.0056439304613777885 | 0.044269530117283296 | 0.0002632173345110476 | 0.00017711867082262535 |
| Go:0031201~snare complex | 13.567708333333334 | 0.038613905348805266 | 0.3709937856060441 | 0.025907305407891434 | 0.09967119503568866 | 0.0002671277845566093 | 1.017019293001842e-05 |
| Go:0008654~phospholipid biosynthetic process | 3.494648544918591 | 0.16125399626286274 | 0.5123341150351401 | 0.40106732181268967 | 0.40106732181268967 | 0.0003048946627728106 | 7.612149882530411e-05 |
| Brain | 1.1520241808378395 | 0.03751473361082591 | 0.38753639661447403 | 0.0060522002534088015 | 0.05316960733436649 | 0.0003175966569555744 | 0.0002750450500793786 |
| Phospholipid biosynthesis | 4.35746827204686 | 0.12679275696108105 | 1.2073963220207706 | 0.05499907359128886 | 0.28781404361562424 | 0.0008744015780815131 | 0.00016614104939798044 |
| Go:0005856~cytoskeleton | 1.4510918003565063 | 0.1017572610526728 | 1.291601191779257 | 0.07081467138361397 | 0.3073533797717233 | 0.0009340046079184327 | 0.0006095581017451592 |
| Amnion | 2.0139208001081226 | 0.062533666777568 | 1.212738685160386 | 0.01702194837244486 | 0.15775545239089095 | 0.0009976698468676399 | 0.00046445637300159226 |
| Go:0016192~vesicle-mediated transport | 1.7715526050585988 | 0.16201760765775464 | 1.6701958399334083 | 0.5684781691905159 | 0.813788909534831 | 0.0009994254964168993 | 0.0005354874549211238 |
| Hippocampus | 1.469091984619935 | 0.11301550671360427 | 1.5766970719435114 | 0.020123941669996204 | 0.2003819081469227 | 0.0012992775935669815 | 0.0008624176811002826 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Ionic channel | 14.828903654485051 | 0.024742508502620053 | 1.864704270884232 | 0.11051722100965067 | 0.11051722100965067 | 0.0018572435401929713 | 0.00012280495398885478 |
| Brain | 1.917745191882305 | 0.49236269120715725 | 8.290172254896454 | 0.3050280540651725 | 0.3050280540651725 | 0.00953018663678688 | 0.004865735606304875 |
| Ion transport | 8.220073664825046 | 0.2093223050914247 | 9.388389919000206 | 0.2641295195986483 | 0.45849463607388385 | 0.009689297868346783 | 0.0011506358417165512 |
| Phosphoprotein | 1.9449572175566472 | 0.7477298884961826 | 12.764140428538406 | 0.24663789925467205 | 0.57242598361481 | 0.013395625849894623 | 0.006728510997732459 |
| Go:0006812~cation transport | 7.035857605177993 | 0.48253718326252226 | 15.907593778513252 | 0.8851640287424223 | 0.8851640287424223 | 0.014420189785834982 | 0.0019944254794716587 |
| Nucleus | 2.3442752100840334 | 0.8823438558218345 | 22.87852190302978 | 0.3324182544375891 | 0.8013823628228709 | 0.02533038885468826 | 0.010435312220344458 |
| Go:0006811~ion transport | 5.0891385767790265 | 0.8805198508203351 | 33.66467718526205 | 0.9229739598893333 | 0.99406698914487 | 0.033825573871062456 | 0.006417516297292802 |
| Go:0016071~mrna metabolic process | 8.998675496688742 | 0.9316052670657596 | 36.66632603116504 | 0.8507133052215513 | 0.996672919503435 | 0.037569112622233884 | 0.004056084295859525 |
| Serine/threonine-protein kinase | 8.7177734375 | 0.6005872417199281 | 34.04773430081717 | 0.40426437679789506 | 0.9249643149402104 | 0.04027433524087503 | 0.004488718566067305 |
| Hippocampus | 4.418547759368398 | 0.7820788913913101 | 37.20070855187346 | 0.6239698075710898 | 0.8586012943818767 | 0.05017564608231821 | 0.010901454302935546 |
| Potassium channel | 31.88214285714286 | 0.5116018538994065 | 44.94799366296351 | 0.4614988118276454 | 0.9756151552300134 | 0.05724524817618929 | 0.0017548905634865639 |
| Go:0030001~metal ion transport | 6.1484162895927605 | 0.9788780757214693 | 60.14985062848338 | 0.9434849223558883 | 0.9999897986578388 | 0.074234364278625 | 0.011621169822539704 |
| Brain cortex | 5.7477338476374165 | 0.863122768651883 | 55.212795612323774 | 0.6756152350024169 | 0.9658664587730105 | 0.0850454754192015 | 0.014205126158360077 |
| Potassium transport | 20.85747663551402 | 0.5623162710130312 | 59.88187930535756 | 0.555948972742046 | 0.9965956817451853 | 0.0862531865080608 | 0.004042096438275098 |
| Spliceosome | 20.105855855855857 | 0.8306086841892719 | 61.23267459299906 | 0.5214413719211177 | 0.9972490594483345 | 0.08933877633615454 | 0.004342367486903085 |
| Potassium | 18.91313559322034 | 0.9502775372925142 | 63.48872753534931 | 0.5016933292140484 | 0.9981056059995418 | 0.0947151697510704 | 0.004892077107615101 |
| Go:0005681~spliceosome | 18.334310850439884 | 0.8323398984486441 | 62.345188999351834 | 0.9954094869006704 | 0.9954094869006704 | 0.09489047780788898 | 0.005060494817242549 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.7038468186859885 | 0.0003352471810086355 | 0.003384080540647094 | 0.0004226498405994761 | 0.0004226498405994761 | 2.837171694763432e-06 | 1.6436427124829718e-06 |
| Brain cortex | 4.029339192158188 | 0.005762231873828294 | 0.18407997876301874 | 0.013769194293189346 | 0.013769194293189346 | 0.00017329584386116217 | 4.157393854157791e-05 |
| Eye | 2.5909920792894696 | 0.014260033074772194 | 0.29286721961993 | 0.010974698152962592 | 0.021828952306376492 | 0.0002758460580717521 | 0.00010332332644701144 |
| Potassium channel | 14.491883116883118 | 0.00990354454306186 | 0.449234296444323 | 0.02773016288704766 | 0.054691363840353224 | 0.0003774039712349808 | 2.4394091785183333e-05 |
| Alternative splicing | 1.6752500456472785 | 0.07118308778862326 | 0.7373603213195135 | 0.030346908118723315 | 0.08830586738381241 | 0.0006202822169670206 | 0.0003619131215604892 |
| Hypothalamus | 4.170639548444279 | 0.04244504803398086 | 1.4296299447063565 | 0.035474403539574983 | 0.10269255287020107 | 0.0013535421996372084 | 0.0003119813128203879 |
| Brain | 1.42842360941749 | 0.1391390604915712 | 1.656439270330945 | 0.030934361684617495 | 0.11811333127791501 | 0.001569912954683525 | 0.0010772775787291522 |
| Potassium transport | 9.480671197960916 | 0.037186849431072866 | 2.1880357399930106 | 0.0667571439814777 | 0.24145963494709388 | 0.0018530411608414594 | 0.00018574703362334816 |
| Potassium | 8.596879815100154 | 0.16418803731891773 | 3.111080324552984 | 0.0759236864846311 | 0.32618730132531915 | 0.002646177929681468 | 0.00029301525907827694 |
| Ionic channel | 4.718287526427062 | 0.13563035478284036 | 4.118063456891086 | 0.0838287294754112 | 0.4086290132661675 | 0.0035193745225302656 | 0.0007142289912687407 |
| Cerebellum | 2.203810518073861 | 0.19609941045410628 | 3.752720710366453 | 0.05594025217453291 | 0.2501102724968408 | 0.0035913993585308082 | 0.0015691249168424506 |
| Coiled coil | 2.1085187906781444 | 0.29917134437704984 | 4.450835943915788 | 0.07803573889613113 | 0.4337625296004185 | 0.0038097828810648676 | 0.0017410896512299776 |
| Voltage-gated channel | 7.514309764309764 | 0.10503843349779995 | 4.995260843360983 | 0.07689768520686502 | 0.4727716923329309 | 0.0042869030256897905 | 0.0005438447298990808 |
| Go:0007017~microtubule-based process | 5.519837508463101 | 0.2190256145434918 | 6.245924893213761 | 0.93569468090581 | 0.93569468090581 | 0.004325708567874795 | 0.0007488495533007334 |
| Go:0005856~cytoskeleton | 2.455542463518535 | 0.27856066687281955 | 5.251176634512378 | 0.4176626983671804 | 0.4176626983671804 | 0.004731799796915698 | 0.0018534336113892815 |
| Go:0043228~non-membrane-bounded organelle | 1.9878997712438504 | 0.5767061669499721 | 5.566725308246323 | 0.24954267789368745 | 0.43681380769702227 | 0.005023698375207577 | 0.0024393174621454277 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.9878997712438504 | 0.5767061669499721 | 5.566725308246323 | 0.24954267789368745 | 0.43681380769702227 | 0.005023698375207577 | 0.0024393174621454277 |
| Go:0007040~lysosome organization | 26.47012987012987 | 0.9649111847053397 | 7.874472946682742 | 0.8253268856722686 | 0.9694893031310513 | 0.00549775108302388 | 0.00018796621832411331 |
| Go:0007010~cytoskeleton organization | 4.168098159509203 | 0.37194851814865804 | 8.759116036691827 | 0.7274881558266986 | 0.9797625332486347 | 0.006142541345658652 | 0.0014085014881617941 |
| Kinase | 2.8696798251253695 | 0.39742676747550587 | 8.527148799904548 | 0.11636171237580295 | 0.6715488941528093 | 0.007444415753074078 | 0.002479988720731657 |
| Go:0006813~potassium ion transport | 6.066071428571428 | 0.6005616932833608 | 12.382559714284858 | 0.7549013954855941 | 0.9963911955111182 | 0.008845917054127562 | 0.0013894821865207659 |
| Serine/threonine-protein kinase | 3.698449337121212 | 0.44412646889671825 | 12.524244922729144 | 0.15392790712486504 | 0.8120364218980902 | 0.011155479232667494 | 0.0028743634013598343 |
| Go:0007033~vacuole organization | 17.127731092436974 | 0.9950355440683867 | 17.489840126253796 | 0.805232057419731 | 0.999719723049757 | 0.012839035244394646 | 0.0006987612292955246 |
| Mmu05014:amyotrophic lateral sclerosis (als) | 15.1 | 0.18841219977766488 | 12.884940404773115 | 0.4591184058096077 | 0.4591184058096077 | 0.014877365598522804 | 0.0009399305239179879 |
| Go:0016071~mrna metabolic process | 3.8565752128666033 | 0.9512122083779179 | 24.298825082818798 | 0.8611060103619851 | 0.9999928203922536 | 0.018537350395352946 | 0.004565719765755441 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mammary tumor | 1.8063880443010407 | 0.009347680730725672 | 0.1407178032770795 | 0.013315006532213514 | 0.013315006532213514 | 0.00012526735006359032 | 6.756350514906849e-05 |
| Cytoplasm | 1.6426889792118806 | 0.029392461882186027 | 0.31454498335710124 | 0.05416941309173928 | 0.05416941309173928 | 0.00024639357628445464 | 0.00014622987398566484 |
| Coiled coil | 1.9152058960865233 | 0.03183032453385959 | 0.3998927819040432 | 0.034796861955450176 | 0.06838290230895372 | 0.0003133729596789931 | 0.00015855567623778132 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.7670220188834225 | 0.21447930013157357 | 1.5585263707472818 | 0.22333137792410518 | 0.22333137792410518 | 0.0012504131425703227 | 0.0006855843942076634 |
| Go:0043228~non-membrane-bounded organelle | 1.7670220188834225 | 0.21447930013157357 | 1.5585263707472818 | 0.22333137792410518 | 0.22333137792410518 | 0.0012504131425703227 | 0.0006855843942076634 |
| Phosphoprotein | 1.3140297147923696 | 0.2205898324115444 | 2.072099127527527 | 0.11607430144429598 | 0.3093670712878629 | 0.0016364772688335712 | 0.001220910164137645 |
| Go:0031201~snare complex | 39.737288135593225 | 0.1577096216466225 | 2.901781226365563 | 0.21093162315412672 | 0.3773710966618189 | 0.0023428186255096844 | 4.432506533989136e-05 |
| Fetus | 14.882397003745318 | 0.01958371329014319 | 2.6261225147211964 | 0.11896824956685492 | 0.2237830547287084 | 0.0023647064812749416 | 0.000142277756372929 |
| Go:0005730~nucleolus | 3.418261344997266 | 0.11525950623148684 | 3.0954140835602773 | 0.1551879299085972 | 0.3970513454353515 | 0.0025014366547410835 | 0.0006955585798219848 |
| Alternative splicing | 1.39344384242881 | 0.3016364467370972 | 3.1650110141491994 | 0.13249699247352753 | 0.43365123678012707 | 0.0025125250425457445 | 0.001758332034777823 |
| Go:0016192~vesicle-mediated transport | 2.799244635193133 | 0.32988044147692575 | 5.216173726541474 | 0.9106983582822807 | 0.9106983582822807 | 0.0035619357476324503 | 0.001212292267063297 |
| Go:0009898~internal side of plasma membrane | 3.322978798795252 | 0.24164887860095197 | 6.698100103761673 | 0.24336832585483248 | 0.6722534935115071 | 0.005507130423597689 | 0.0015704064133956264 |
| Go:0005856~cytoskeleton | 1.8888788180911809 | 0.506495740864003 | 9.486070813032544 | 0.2743783162628898 | 0.7988355374378 | 0.007907345824376995 | 0.004004595305399631 |
| Go:0031981~nuclear lumen | 2.0401174731750387 | 0.4839330613644881 | 9.61593871318679 | 0.237476883171722 | 0.80342961586986 | 0.008020802512980272 | 0.00375157471565239 |
| Lens | 17.62389118864577 | 0.3485648820885029 | 12.90489293254401 | 0.3549416740733158 | 0.7315910732861521 | 0.012216758408779019 | 0.0006164681444550803 |
| Activated spleen | 2.2427105222027333 | 0.5440904440822573 | 14.006496322178974 | 0.3016974505284499 | 0.7622204446767411 | 0.013334698963269973 | 0.005634861623697406 |
| Go:0005829~cytosol | 2.3161989441511532 | 0.6291490455613262 | 15.948819303383077 | 0.3292531779765049 | 0.9389184054733029 | 0.013744006879247522 | 0.005620255105176232 |
| Tpr repeat | 4.151610277874666 | 0.4950433295684926 | 17.366050910992147 | 0.4905385297044592 | 0.965679253683977 | 0.014809619953749202 | 0.003343821711287184 |
| Skeletal muscle | 4.034384729930959 | 0.659538477789473 | 17.109549180815854 | 0.3004030834251211 | 0.8324133446791445 | 0.016555397183308077 | 0.003868229853241864 |
| Visual cortex | 3.36778380472685 | 0.5016694666639804 | 18.360081619495162 | 0.27517606123252614 | 0.8549909290420818 | 0.0178844854383243 | 0.004998198884390214 |
| Corpus striatum | 4.72957531898686 | 0.444981229245622 | 21.510895807394505 | 0.2806257851884064 | 0.9003025458010371 | 0.021317309593064198 | 0.004226677920094634 |
| Salivary gland | 2.2323595505617977 | 0.7821488203437883 | 25.506709136597138 | 0.2955755175612893 | 0.9393716802321048 | 0.0258560541095786 | 0.010903738069312326 |
| Embryo | 1.513464102075795 | 0.9087108766101724 | 26.558587210508588 | 0.27854476418252416 | 0.9470477393189088 | 0.02708771568813188 | 0.017073598845370304 |
| Brain | 1.2057733111881719 | 0.9562327616036139 | 26.96259907394548 | 0.2585054601564182 | 0.9497565360398045 | 0.027565055688740108 | 0.0222583976415422 |
| Liver | 1.3588275525158768 | 0.94396107107911 | 28.17784855675799 | 0.24905095949385936 | 0.9571732484493867 | 0.02901551401950541 | 0.020518292438443213 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0009898~internal side of plasma membrane | 13.070383275261324 | 0.20097437518147765 | 13.689027458695035 | 0.4121117022628775 | 0.4121117022628775 | 0.016990087348873018 | 0.0012739733890769658 |
| Embryo | 3.0066585956416465 | 0.9379909501010342 | 43.29003061891302 | 0.8884195890203911 | 0.8884195890203911 | 0.06246415523262538 | 0.01980467585345658 |
| Protein transport | 6.8345029985964025 | 0.8379753226468493 | 47.722315071369195 | 0.9783375743264836 | 0.9783375743264836 | 0.06288768424544078 | 0.008881921713649514 |
| Go:0007611~learning or memory | 25.44569288389513 | 0.98811722337422 | 50.897206391599696 | 0.9812617940624647 | 0.9812617940624647 | 0.06975990448134603 | 0.0026828584851511512 |
| Go:0015031~protein transport | 5.218125960061444 | 0.9969611314585858 | 62.4007305719651 | 0.9351000526426015 | 0.9957879968330069 | 0.09466588625865463 | 0.017411098765985303 |
| Go:0045184~establishment of protein localization | 5.178353658536586 | 0.9973072654993678 | 62.908006719981266 | 0.8425411339035209 | 0.9960960759592832 | 0.09591530840834792 | 0.017771101120698938 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mammary tumor | 1.7669092932924322 | 3.127689901856058e-51 | 5.818500892244427e-50 | 1.1046091290245013e-50 | 1.1046091290245013e-50 | 4.4902810122947205e-53 | 2.2501366200403295e-53 |
| Bone marrow | 2.0916124235871245 | 1.445028028558914e-48 | 3.3658120872360227e-47 | 3.194900909182277e-48 | 6.389801818364553e-48 | 2.5974804139693305e-50 | 1.0395885097546144e-50 |
| Acetylation | 1.669913056958673 | 2.0070080396431114e-37 | 2.70779318665187e-36 | 1.06515151763417e-36 | 1.06515151763417e-36 | 1.8428226948688062e-39 | 9.838274704132899e-40 |
| Kidney | 1.7140812142191453 | 1.7836107485549052e-35 | 3.1900043117471377e-34 | 2.0186802692770438e-35 | 6.056040807831131e-35 | 2.4618052064354193e-37 | 1.2831732003992123e-37 |
| Go:0005739~mitochondrion | 1.8778480980346441 | 6.218875603334512e-35 | 1.1315739429135476e-33 | 4.251373444478029e-34 | 4.251373444478029e-34 | 7.757980738098593e-37 | 3.533452047349154e-37 |
| Hippocampus | 1.993891001452438 | 1.285216942129677e-29 | 2.803720374329414e-28 | 1.330675709289795e-29 | 5.32270283715918e-29 | 2.1637003403086095e-31 | 9.246165051292642e-32 |
| Mitochondrion | 2.1088506207448936 | 6.326115942632376e-29 | 1.1702449127814607e-27 | 2.301667924635885e-28 | 4.60333584927177e-28 | 7.964248874172613e-31 | 3.101037226780576e-31 |
| Ribonucleoprotein | 2.822439559049807 | 6.291488819251587e-23 | 1.806531781064506e-21 | 2.3687555826404516e-22 | 7.106266747921355e-22 | 1.2294579148652864e-24 | 3.084063146691954e-25 |
| Transit peptide | 2.3312403768548515 | 1.0489650764664275e-22 | 2.2408186523568347e-21 | 2.2036500055346554e-22 | 8.814600022138622e-22 | 1.5250173048682736e-24 | 5.141985668953076e-25 |
| Ribosomal protein | 3.2317198717557116 | 2.0791409427899032e-21 | 3.759548907444587e-20 | 2.957751163659552e-21 | 1.478875581829776e-20 | 2.5586082730618963e-23 | 5.095933683308586e-24 |
| Go:0005840~ribosome | 2.9302587176602923 | 1.046311423699294e-18 | 3.7037409899769784e-17 | 6.95755950754333e-18 | 1.391511901508666e-17 | 2.539255294723843e-20 | 5.944951271018716e-21 |
| Liver | 1.3629997202171116 | 5.752962226153164e-17 | 7.728251207383378e-16 | 2.934328615966702e-17 | 1.4671643079833507e-16 | 5.964082552777849e-19 | 4.1388217454339306e-19 |
| Mammary gland | 1.4807567858415316 | 2.2499725642545086e-16 | 3.345327027776419e-15 | 1.0584853506910832e-16 | 6.3509121041465e-16 | 2.5816715870514225e-18 | 1.6186852980248263e-18 |
| Go:0044429~mitochondrial part | 1.999733811899467 | 4.767445025890368e-16 | 9.446092951235093e-15 | 1.1829796593922955e-15 | 3.5489389781768867e-15 | 6.476166018570961e-18 | 2.7087755828922546e-18 |
| Go:0031090~organelle membrane | 1.7733679458176503 | 7.045989141162987e-16 | 1.185953361135171e-14 | 1.1139198320831103e-15 | 4.455679328332441e-15 | 8.130801694037302e-18 | 4.003402921115334e-18 |
| Cytoplasm | 1.3573303480849188 | 1.907267244511698e-15 | 1.978999985476503e-14 | 1.2974494312191269e-15 | 7.784696587314761e-15 | 1.34683331960463e-17 | 9.349349237802441e-18 |
| Go:0006412~translation | 2.3572557435122556 | 9.956364670210849e-15 | 8.17799386116679e-14 | 1.5460375345592517e-13 | 1.5460375345592517e-13 | 4.470900909656598e-17 | 1.5085401015470982e-17 |
| Go:0030529~ribonucleoprotein complex | 2.0245423867471115 | 1.9539925233402755e-14 | 3.219646771412954e-13 | 2.4313884239290928e-14 | 1.2168044349891716e-13 | 2.6582390048637366e-16 | 6.366360313667965e-17 |
| Embryo | 1.4524831022651845 | 9.259260025373806e-14 | 1.2989609388114332e-12 | 3.5083047578154947e-14 | 2.4580337765200966e-13 | 9.855947838350597e-16 | 7.205130480503212e-16 |
| Go:0019866~organelle inner membrane | 2.1864238124080644 | 1.6413537196058314e-12 | 3.7247982476174e-11 | 2.3322455078300663e-12 | 1.3993251002375473e-11 | 2.5483400860238986e-14 | 9.306205289750673e-15 |
| Go:0005743~mitochondrial inner membrane | 2.2095734654789774 | 2.9114488597770105e-12 | 6.687983500341943e-11 | 3.589573083218056e-12 | 2.5127011582526393e-11 | 4.582766236243274e-14 | 1.651154625647078e-14 |
| Go:0006091~generation of precursor metabolites and energy | 2.362494089608949 | 1.527777904186678e-11 | 8.488765246283947e-11 | 8.023826048031424e-11 | 1.6047652096062848e-10 | 4.641163658133514e-14 | 1.5464211832791026e-14 |
| Tongue | 1.826438683581541 | 7.299383320003017e-12 | 1.4142020887675244e-10 | 3.3558711365344607e-12 | 2.684719113688061e-11 | 1.0910061380386044e-13 | 5.24690844659042e-14 |
| Colon | 1.6196058959974506 | 9.398148925754413e-12 | 1.527777904186678e-10 | 3.2227553958819044e-12 | 2.900479856293714e-11 | 1.178975405707954e-13 | 6.76540385629798e-14 |
| Go:0005740~mitochondrial envelope | 1.978486704507205 | 4.929923136387515e-11 | 9.69679891937858e-10 | 4.5538905979469746e-11 | 3.6431124783575797e-10 | 6.648048063598619e-13 | 2.8006058300922894e-13 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Bone marrow | 2.2531036708398804 | 3.428234430391604e-25 | 7.50499075715267e-24 | 1.1624143642511983e-24 | 1.1624143642511983e-24 | 6.022872353633152e-27 | 2.466355705317701e-27 |
| Mammary tumor | 1.8421227593804363 | 6.396024173922419e-25 | 1.1142963557092561e-23 | 8.629418288588393e-25 | 1.7258836577176787e-24 | 8.94240237159419e-27 | 4.601456240231956e-27 |
| Hippocampus | 2.385636515573152 | 8.373848685883306e-22 | 1.9579821053472573e-20 | 1.0108768941668646e-21 | 3.032630682500594e-21 | 1.5713112344562664e-23 | 6.024351572577918e-24 |
| Acetylation | 1.7207211293157207 | 1.3810453385616182e-17 | 1.7526708070860493e-16 | 5.865289382723382e-17 | 5.865289382723382e-17 | 1.2244863011948605e-19 | 6.769830090988325e-20 |
| Kidney | 1.7642148882640225 | 1.9491535834595426e-16 | 3.2352778194795155e-15 | 1.2527441843023253e-16 | 5.010976737209301e-16 | 2.596361003735389e-18 | 1.4022687650788076e-18 |
| Go:0005739~mitochondrion | 1.8273208052161476 | 1.270095140171179e-12 | 1.9817480989559044e-11 | 6.225020499073253e-12 | 6.225020499073253e-12 | 1.3978817733315477e-14 | 7.219171983756373e-15 |
| Cytoplasm | 1.4709988563975303 | 2.0150414670183636e-10 | 2.0804802325358196e-09 | 3.4811420412950156e-10 | 6.962284082590031e-10 | 1.4534967252869117e-12 | 9.878185218835808e-13 |
| Mitochondrion | 2.065377855887522 | 1.7335199942181134e-10 | 2.698963275094002e-09 | 3.0106794834949824e-10 | 9.032037340261923e-10 | 1.8855615937983834e-12 | 8.498114012184598e-13 |
| Mammary gland | 1.5656991828520161 | 9.52949152921434e-09 | 1.392638004915625e-07 | 4.313989165893872e-09 | 2.156994571844706e-08 | 1.1176142859692829e-10 | 6.855750482485125e-11 |
| Visual cortex | 3.087376054462039 | 7.658179645986252e-09 | 2.4490240946306585e-07 | 6.321972612077786e-09 | 3.79318348953106e-08 | 1.965379551716325e-10 | 5.509482630148921e-11 |
| Ribonucleoprotein | 2.713122877492501 | 1.0474786094061272e-07 | 2.272288013305257e-06 | 1.901045187935324e-07 | 7.604178582365506e-07 | 1.5875118059380864e-09 | 5.134699377528251e-10 |
| Transit peptide | 2.2314667753708184 | 1.716050341826758e-07 | 2.984676117367968e-06 | 1.9976355603024132e-07 | 9.988173811370515e-07 | 2.0852148041248056e-09 | 8.412012329156626e-10 |
| Embryo | 1.519588481642748 | 3.948083657778767e-07 | 5.665621183315039e-06 | 1.253603754136634e-07 | 8.775222981594055e-07 | 4.546749626921669e-09 | 2.8403484598488577e-09 |
| Go:0006091~generation of precursor metabolites and energy | 2.713498200394752 | 1.7602378796777174e-06 | 9.626245245364373e-06 | 1.3089164913937523e-05 | 1.3089164913937523e-05 | 5.485855203436075e-09 | 1.7780196751350227e-09 |
| Liver | 1.3692693910555453 | 8.766458176046044e-07 | 1.1285471057664864e-05 | 2.1849448905708613e-07 | 1.7479545760812343e-06 | 9.056767333672317e-09 | 6.3068071350219325e-09 |
| Go:0031090~organelle membrane | 1.8029008406339972 | 1.230565509380277e-06 | 1.9516324489821102e-05 | 3.065714686001364e-06 | 6.131419973409713e-06 | 1.377851395343636e-08 | 6.991853768109858e-09 |
| Brain | 1.2009913558604848 | 7.7799081286134e-06 | 8.530227010306035e-05 | 1.4680091153795871e-06 | 1.3212004456919502e-05 | 6.845643149073696e-08 | 5.597077826265496e-08 |
| Ribosomal protein | 2.836416351414415 | 3.0758781828232884e-05 | 0.00035277640172415303 | 1.9675866195556502e-05 | 0.00011804939022974636 | 2.4646418380211816e-07 | 7.5390327784808e-08 |
| Salivary gland | 1.9844744440109245 | 1.7158027268759923e-05 | 0.0003311861207389022 | 5.129588650509831e-06 | 5.129470245091028e-05 | 2.6578244172383285e-07 | 1.2344009643888915e-07 |
| Tongue | 1.867827508685878 | 5.5053545743311005e-05 | 0.001001530172994869 | 1.4102018220718548e-05 | 0.00015511126321021695 | 8.037473153474436e-07 | 3.960794988224189e-07 |
| Go:0019866~organelle inner membrane | 2.1913262921768393 | 8.621734249447677e-05 | 0.0017106684723544596 | 0.00017913223043286575 | 0.0005373004319786379 | 1.2077404607948982e-06 | 4.898922630863753e-07 |
| Go:0008610~lipid biosynthetic process | 2.3116214779372677 | 0.00038555291313790985 | 0.0026419177326486576 | 0.0017945802771035524 | 0.0035859400358361215 | 1.5056086447770384e-06 | 5.842835532286489e-07 |
| Gtp-binding | 2.205472298803461 | 0.00012980488526270761 | 0.0022510413913767735 | 0.00010761084748200922 | 0.0007530327940001857 | 1.5726844953346974e-06 | 6.36339555577323e-07 |
| Go:0006412~translation | 2.22013489123207 | 0.00043899834682514793 | 0.002897090340558961 | 0.0013122597374123357 | 0.003931615395126653 | 1.6510315425663254e-06 | 6.652948314783879e-07 |
| Cytoplasmic vesicle | 2.4344888188594633 | 0.00012682298608945874 | 0.00243008919964538 | 0.00010164933052003722 | 0.0008129053905507888 | 1.6977772170707005e-06 | 6.21720536118442e-07 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Hippocampus | 2.915967077222933 | 0.0001934206737568278 | 0.004735438420744309 | 0.00046436264300853125 | 0.00046436264300853125 | 4.184410098775766e-06 | 1.391649261549905e-06 |
| Mammary tumor | 1.8113456310180305 | 0.018609030037795016 | 0.2842086828480195 | 0.01386071194899019 | 0.027529304562247447 | 0.0002514579032101311 | 0.00013513012484605918 |
| Brain | 1.3499655595030717 | 0.07233593889044554 | 0.8368302006633299 | 0.027100352931517024 | 0.0791176746961122 | 0.0007422741015090948 | 0.0005400384062328109 |
| Acetylation | 1.7496774193548386 | 0.09660801819141718 | 1.135550500199467 | 0.17788225271707792 | 0.17788225271707792 | 0.0008980902795020346 | 0.0004979089990776869 |
| Bone marrow | 2.081299677437659 | 0.06602151784457078 | 1.192576361548059 | 0.02898972125975008 | 0.11101320743162835 | 0.001059554492274618 | 0.0004912597197203282 |
| Kidney | 1.8155950596130506 | 0.07951837955627772 | 1.2569835798582174 | 0.02450835045738453 | 0.11668057642240093 | 0.0011171088632917468 | 0.0005959248513575596 |
| Mammary tumor metastatized to lung. tumor arose spontaneously | 3.3517061575032594 | 0.06432997475307145 | 1.8839718303491293 | 0.030612616777789592 | 0.17017946474976975 | 0.0016791816048034162 | 0.0004782482056556851 |
| Mmu00270:cysteine and methionine metabolism | 12.64573002754821 | 0.10569505194510165 | 3.5745109398316655 | 0.21604285496416387 | 0.21604285496416387 | 0.0035213351016007387 | 0.000251563975819844 |
| Cytoplasm | 1.5295477055133706 | 0.4292970082110187 | 5.373594603622012 | 0.3772772430387267 | 0.6122163679625509 | 0.0043360205684966294 | 0.002745666680382018 |
| Placenta | 2.4196809158445984 | 0.24850754990520973 | 5.736719028894221 | 0.07944518422264701 | 0.4397942267732585 | 0.005206678291222107 | 0.0020532425622653066 |
| Go:0008652~cellular amino acid biosynthetic process | 10.741501976284585 | 0.6351204684041767 | 8.597066973550948 | 0.9853622510882408 | 0.9853622510882408 | 0.005931845935171426 | 0.0005090562607621902 |
| Go:0005829~cytosol | 2.42694616142614 | 0.3769377451558771 | 8.223656230359955 | 0.7383258683774376 | 0.7383258683774376 | 0.006851575684001075 | 0.002684508640795805 |
| Mmu00900:terpenoid backbone biosynthesis | 22.355844155844153 | 0.06059610001969917 | 7.337808942041013 | 0.22493127439423777 | 0.39926847058785964 | 0.007358404276282377 | 0.00028153582906345674 |
| Heart | 2.1207944439626125 | 0.40539830122534837 | 9.003347522645699 | 0.10923257262793862 | 0.6036173628659918 | 0.008302060189091631 | 0.0037330400784881816 |
| Cholesterol biosynthesis | 18.833333333333336 | 0.4015762481959082 | 12.800954692970167 | 0.5430096432898728 | 0.9045620488468473 | 0.010718650481999832 | 0.0005032617143822897 |
| Amnion | 3.2689728929291264 | 0.3671325490598132 | 12.124642456183832 | 0.13138643975682762 | 0.718525248052474 | 0.011355863425766133 | 0.0032859152585150322 |
| Amino-acid biosynthesis | 16.142857142857142 | 0.8356743135056635 | 16.901981363547193 | 0.5479084551013778 | 0.9582260503055792 | 0.014460855550929883 | 0.000804446725756898 |
| Liver | 1.4307678458907194 | 0.756673533869205 | 15.733477094095994 | 0.15456292613914768 | 0.8134424263771707 | 0.015012437408839532 | 0.010116475348649196 |
| Go:0006695~cholesterol biosynthetic process | 15.411720226843098 | 0.9999995652900862 | 21.3093152599092 | 0.9964135720608984 | 0.9999871375346376 | 0.01573572029668517 | 0.0009243570446497681 |
| Go:0051186~cofactor metabolic process | 3.8952699474438606 | 0.9883328084618278 | 24.652475175745202 | 0.988130123306188 | 0.9999983276059169 | 0.018559767433892295 | 0.004485842275490494 |
| Sterol biosynthesis | 14.125 | 0.8592263464311881 | 21.314280087334993 | 0.5605557944852853 | 0.9836122733467629 | 0.018682104351876744 | 0.0012006279166153872 |
| Gtp-binding | 2.888178913738019 | 0.7518632241800212 | 23.540818786880312 | 0.535714544144485 | 0.9899836273114472 | 0.020895724253670082 | 0.00680894459440853 |
| Go:0009309~amine biosynthetic process | 6.474329958308516 | 0.9987796376480579 | 30.069086368025545 | 0.9850304285561826 | 0.9999999497845372 | 0.023393772315781654 | 0.0033824521594607686 |
| Go:0016126~sterol biosynthetic process | 11.815652173913042 | 0.9999999997542717 | 32.8855598303572 | 0.9764289480319613 | 0.9999999927239642 | 0.02604726826069132 | 0.0020297824237214476 |
| Medulla oblongata | 2.3416266927528375 | 0.7822756890986516 | 26.751983798163394 | 0.2423910767031946 | 0.9528050105078291 | 0.027133797606314874 | 0.010907883256063056 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Mammary gland | 1.6507145230973748 | 0.004096443369772995 | 0.05978335397809342 | 0.007318807169880537 | 0.007318807169880537 | 5.031189210714711e-05 | 2.953090768417232e-05 |
| Bone marrow | 1.7919211126602306 | 0.00844027131186531 | 0.13389655127868938 | 0.008195376407871602 | 0.016323588621276475 | 0.00011272163436958687 | 6.0977220686383836e-05 |
| Go:0001944~vasculature development | 2.767010909090909 | 0.3186243179148083 | 2.8269266942151927 | 0.9322401076083284 | 0.9322401076083284 | 0.0017152268147119505 | 0.0005811060357155166 |
| Liver | 1.3396233434085099 | 0.2461950225794678 | 3.274652297422409 | 0.12744168769780284 | 0.33567074116095363 | 0.002797295194362396 | 0.002031183235082312 |
| Mammary tumor. wap-tag model. 5 months old | 4.767448238036473 | 0.16869416400321713 | 4.04672869356093 | 0.11913862248405827 | 0.3979531716204815 | 0.0034694472963188827 | 0.0006643745944074048 |
| Go:0001568~blood vessel development | 2.632548435171386 | 0.759884944843044 | 6.509614856602508 | 0.9575391805135927 | 0.9981970788085427 | 0.0040214502894703515 | 0.0014400097390731407 |
| Go:0006081~cellular aldehyde metabolic process | 11.626096256684493 | 0.995685331721741 | 7.214311484761026 | 0.9039469420661163 | 0.9991137962433331 | 0.004472470914898446 | 0.00031730016444307403 |
| Mammary tumor | 1.3795583857794664 | 0.35792441507468264 | 5.284162147227089 | 0.12486082623512873 | 0.486683029143939 | 0.004557130841547449 | 0.003182330285406714 |
| Go:0001525~angiogenesis | 3.3435953520164046 | 0.8673234405252322 | 8.844425148979962 | 0.8861739459821165 | 0.9998321324615678 | 0.005528243926821557 | 0.0015290124404711833 |
| Oxidoreductase | 1.9021947581504368 | 0.47179355232873443 | 8.177439793364439 | 0.8739750518362938 | 0.8739750518362938 | 0.006275893673063479 | 0.003123875575663691 |
| Lung | 1.5461170085401867 | 0.4694499815531673 | 8.39551666096362 | 0.1643322654621696 | 0.6594335947861074 | 0.007350558034026635 | 0.004549626432346572 |
| Mammary tumor. mmtv-ltr/int3 model. 5 month old mouse. taken by biopsy. | 2.7306212204507974 | 0.29655307003353826 | 8.426165328888313 | 0.1431265940904487 | 0.6608306107679378 | 0.007378504620897247 | 0.0025274684904747105 |
| Cytoskeleton | 1.8663042910155228 | 0.548470637766924 | 9.911135249308089 | 0.7183336621706569 | 0.9206640741338064 | 0.0076727359662468915 | 0.0038900360696828952 |
| Go:0000272~polysaccharide catabolic process | 9.411601731601731 | 0.9999165553466145 | 12.913788441234708 | 0.9254162519519925 | 0.9999976920785656 | 0.008243363182329693 | 0.0007486292846579668 |
| Kidney | 1.3921888253734096 | 0.566811453374916 | 9.999813911802601 | 0.14936497663796855 | 0.7258766186245156 | 0.008825054446325538 | 0.006000501647730409 |
| Go:0000902~cell morphogenesis | 2.238681965283907 | 0.7467811117007414 | 15.337306378920768 | 0.9260774499588957 | 0.9999998368219873 | 0.009917600229330362 | 0.0041534745871125035 |
| Go:0032989~cellular component morphogenesis | 2.1115773115773115 | 0.8276097088549146 | 18.225074410443153 | 0.9326601938089455 | 0.9999999937208185 | 0.011972413008974406 | 0.005313090607020438 |
| Go:0016324~apical plasma membrane | 3.6383588976181573 | 0.6692313380658312 | 15.26864424862745 | 0.9719025824013344 | 0.9719025824013344 | 0.012412108230136533 | 0.0031380661004337808 |
| Go:0045765~regulation of angiogenesis | 5.49010101010101 | 0.9997200603083101 | 19.093450788449307 | 0.9167592010424109 | 0.9999999976948992 | 0.012603663753075301 | 0.002063760533180493 |
| Cell shape | 7.913130193905817 | 0.7027709115978316 | 16.77434448147883 | 0.7737192911079416 | 0.9884137580910757 | 0.013458572784413965 | 0.001485723767169605 |
| Lung and heart | 7.870073281837988 | 0.19275579407590648 | 15.173548706322826 | 0.20116580744640133 | 0.8675323562003003 | 0.013749915324998392 | 0.0015393107843779852 |
| Go:0007010~cytoskeleton organization | 2.121940881204685 | 0.8885096438013746 | 22.330417851323446 | 0.9283275367437335 | 0.9999999999500919 | 0.01501423079474215 | 0.006625882252438197 |
| Mmu00010:glycolysis / gluconeogenesis | 4.05035294117647 | 0.6827165137869975 | 16.631921459248446 | 0.8830085270663757 | 0.8830085270663757 | 0.015317831398962057 | 0.0034413909987555573 |
| Go:0007548~sex differentiation | 3.0406713286713285 | 0.9999889813277635 | 23.93291090826981 | 0.9232942064040351 | 0.9999999999929485 | 0.016242775504061856 | 0.004929765009400627 |
| Go:0045177~apical part of cell | 3.0268700072621644 | 0.5901104427885597 | 19.860838928030933 | 0.9080673350222652 | 0.9915483851100916 | 0.016551692140417427 | 0.005054611378784068 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Glycoprotein | 1.8597916666666665 | 0.09707534931126138 | 1.137112669012208 | 0.13518046673247885 | 0.13518046673247885 | 0.0009550333090545902 | 0.0005004442083533052 |
| Go:0007218~neuropeptide signaling pathway | 11.810517166449369 | 0.5646186116003884 | 6.333422063355698 | 0.9144100344752808 | 0.9144100344752808 | 0.004475697197673978 | 0.00035990616069121327 |
| Mammary gland | 2.0502025127054146 | 0.41900364968330694 | 8.304042502547393 | 0.44734779526346313 | 0.44734779526346313 | 0.008317699420791108 | 0.0038989318961986977 |
| Go:0006887~exocytosis | 8.374730354391371 | 0.5801263579429721 | 15.56857927748636 | 0.9583741435441901 | 0.9982672880743203 | 0.011535265694333498 | 0.0013139866674181436 |
| Go:0005886~plasma membrane | 1.6733195687084192 | 0.7182551045324621 | 13.437662095263182 | 0.7854775297604575 | 0.7854775297604575 | 0.0124369792579763 | 0.007171621914360452 |
| Disulfide bond | 1.8078169299311462 | 0.8054340866151867 | 16.618059001766195 | 0.6846239503282786 | 0.9005379472934599 | 0.015069376163052497 | 0.00799232296395646 |
| Membrane | 1.4409135847305448 | 0.8770589038500688 | 16.750637188054775 | 0.5397854457758854 | 0.9025277374424046 | 0.015200313600655331 | 0.010222149596894665 |
| Go:0043085~positive regulation of catalytic activity | 4.411974803558672 | 0.995362874586479 | 30.999747448472846 | 0.9904093166787239 | 0.9999991178373763 | 0.025118719734796163 | 0.005413235378829521 |
| Whole body | 2.200197118525819 | 0.8719043930232958 | 30.260836591377682 | 0.7084978214900397 | 0.9150264799239473 | 0.034128197664399354 | 0.014675269995869305 |
| Carbohydrate metabolism | 10.190639269406391 | 0.7290464962869192 | 34.04915998129901 | 0.7332839314849185 | 0.9949394616134293 | 0.03418036329675511 | 0.0031953937439811774 |
| Signal | 1.5863542835765059 | 0.9928294886459521 | 38.5248438007646 | 0.7093836012518435 | 0.9979269938813039 | 0.03983458992089478 | 0.023914208816378815 |
| Go:0044093~positive regulation of molecular function | 3.7631549795059267 | 0.9683193260981122 | 46.03795558995118 | 0.9969548368069744 | 0.9999999999140111 | 0.04141154543714738 | 0.010406229457914522 |
| Go:0032940~secretion by cell | 4.952797521414252 | 0.9999889999381717 | 48.91078588327803 | 0.9935677942632848 | 0.9999999999889897 | 0.04500019258089988 | 0.008612403224276845 |
| Go:0016192~vesicle-mediated transport | 2.9653015203317086 | 0.993828918371172 | 51.329450108026656 | 0.9889911380613516 | 0.9999999999982199 | 0.048170362148280264 | 0.015299575539163906 |
| Go:0016010~dystrophin-associated glycoprotein complex | 38.592592592592595 | 0.9448314611210168 | 44.456082858656366 | 0.9565502388550992 | 0.9981121182564511 | 0.04971592019598723 | 0.0011751865388432016 |
| Go:0035023~regulation of rho protein signal transduction | 8.033898305084746 | 0.9978059524141079 | 54.173324526124404 | 0.9848241741890071 | 0.9999999999998146 | 0.05209122335276477 | 0.006164765062185777 |
| Egf-like domain | 4.467967967967968 | 0.9938132215428247 | 51.4402082707842 | 0.7832390300761314 | 0.9998962742666777 | 0.05856849675813946 | 0.012386715708708713 |
| Surface antigen | 30.99652777777778 | 0.8970110153833009 | 53.391846335351815 | 0.7496653483859212 | 0.9999383906275348 | 0.06178945950603582 | 0.0018554115934106515 |
| Glucose metabolism | 29.173202614379083 | 0.8825236240519153 | 55.56441770376905 | 0.7240713726298922 | 0.9999663973070047 | 0.06552378781516394 | 0.0020973250057809826 |
| Go:0006904~vesicle docking during exocytosis | 28.788135593220336 | 1.0 | 63.16932621453306 | 0.9908211534933058 | 1.0 | 0.06618750578974904 | 0.0021388735207519363 |
| Mmu04080:neuroactive ligand-receptor interaction | 4.012587412587413 | 0.8293607372410627 | 47.88030665877374 | 0.955756772951231 | 0.955756772951231 | 0.06694400721144231 | 0.015803553879459368 |
| Go:0046903~secretion | 4.168417823452719 | 0.9940975008098101 | 64.35212174760497 | 0.9865113820598715 | 1.0 | 0.06827491840421582 | 0.015432346798141655 |
| Go:0030198~extracellular matrix organization | 6.840745091458299 | 0.9999999999996273 | 64.83236895627711 | 0.9802830574899501 | 1.0 | 0.06914093208867438 | 0.009589368731320956 |
| Go:0006081~cellular aldehyde metabolic process | 27.094715852442672 | 1.0 | 65.3996290563839 | 0.9733478221964047 | 1.0 | 0.07017815747458123 | 0.002417304798681189 |
| Go:0048278~vesicle docking | 27.094715852442672 | 1.0 | 65.3996290563839 | 0.9733478221964047 | 1.0 | 0.07017815747458123 | 0.002417304798681189 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0006915~apoptosis | 19.481003584229388 | 0.6778179692320478 | 53.19835403368811 | 0.9982314455833609 | 0.9982314455833609 | 0.06727407379968021 | 0.003426354725622144 |
| Go:0012501~programmed cell death | 19.151515151515152 | 0.6901253384511872 | 53.81653515322966 | 0.9602160033318389 | 0.9984172336091077 | 0.06841098108519049 | 0.0035439700519336865 |
| Go:0008219~cell death | 17.867192636423404 | 0.739281936987189 | 56.357960720225975 | 0.9004417761270284 | 0.9990131948201867 | 0.07323510223808677 | 0.004065397665675466 |
| Go:0016265~death | 17.45407835581246 | 0.7553813081382847 | 57.22264154850081 | 0.8300107642851349 | 0.9991650015196405 | 0.07493474331201534 | 0.004257742248547163 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.332756979136204 | 9.10148476751409e-62 | 9.7096525313432e-61 | 4.114303852346101e-61 | 4.114303852346101e-61 | 6.530641035470001e-64 | 4.46151214093828e-64 |
| Brain | 1.2898884444885164 | 1.3371241912263749e-50 | 1.785440425061855e-49 | 4.030953972027384e-50 | 4.030953972027384e-50 | 1.3347529708699946e-52 | 9.619598498031474e-53 |
| Acetylation | 1.5686603836232216 | 3.445215354072751e-46 | 4.648676596614794e-45 | 9.848997154132749e-46 | 1.9697994308265497e-45 | 3.126665763216746e-48 | 1.6888310559180152e-48 |
| Embryo | 1.500389178761781 | 4.681730164927453e-28 | 7.691504845246816e-27 | 8.682480124124392e-28 | 1.7364960248248784e-27 | 5.749986837168471e-30 | 3.368151197789534e-30 |
| Mammary tumor | 1.423939443891208 | 2.826647325592346e-27 | 4.3165811830581554e-26 | 3.2484870065648054e-27 | 9.745461019694416e-27 | 3.2269738475809327e-29 | 2.0335592270448532e-29 |
| Bone marrow | 1.5988453786029022 | 3.690970932811386e-26 | 6.640535681270782e-25 | 3.748051923922453e-26 | 1.4992207695689813e-25 | 4.964307184003249e-28 | 2.6553747718067527e-28 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.4451691327374405 | 2.3202107264499207e-23 | 1.605639481338055e-22 | 6.943421768928944e-23 | 6.943421768928944e-23 | 1.076499499058751e-25 | 6.591507745596366e-26 |
| Go:0043228~non-membrane-bounded organelle | 1.4451691327374405 | 2.3202107264499207e-23 | 1.605639481338055e-22 | 6.943421768928944e-23 | 6.943421768928944e-23 | 1.076499499058751e-25 | 6.591507745596366e-26 |
| Cytoplasm | 1.3300800104139792 | 3.6820893321960506e-22 | 3.916200245545599e-21 | 5.531415844454893e-22 | 1.659424753336468e-21 | 2.6340075449785205e-24 | 1.8049457510764954e-24 |
| Mmu04360:axon guidance | 2.865153629374265 | 1.100659471931285e-18 | 6.288753212873992e-17 | 9.402260340788736e-18 | 9.402260340788736e-18 | 5.0823028869128305e-20 | 9.915851098480044e-21 |
| Go:0007049~cell cycle | 1.7443877527094012 | 4.3685431707728193e-17 | 5.326401065358898e-16 | 1.1921593657411255e-15 | 1.1921593657411255e-15 | 2.852738372196998e-19 | 1.3238009608402483e-19 |
| Mammary gland | 1.3600199857908974 | 1.904691993682446e-15 | 2.7304471580026614e-14 | 1.2328954427678991e-15 | 6.1644772138394965e-15 | 2.0412176204766544e-17 | 1.3702820098434864e-17 |
| Go:0051301~cell division | 2.0783326025553435 | 7.148465315313469e-15 | 5.792882485773478e-14 | 6.482838060174547e-14 | 1.2965676120349095e-13 | 3.102578636120865e-17 | 1.0831008053505256e-17 |
| Cell cycle | 1.8131421287218448 | 3.463895692470493e-15 | 5.840471198477806e-14 | 6.187006454104678e-15 | 2.474802581641871e-14 | 3.928258066098208e-17 | 1.6979880845443594e-17 |
| Go:0022402~cell cycle process | 1.8699264346697582 | 3.6637359812630166e-14 | 6.217248937900877e-13 | 4.639622019908529e-13 | 1.3918866059725588e-12 | 2.8773463073871405e-16 | 7.353743675060512e-17 |
| Cerebellum | 1.4140856173084815 | 4.629630012686903e-14 | 5.995204332975845e-13 | 2.2315482794965646e-14 | 1.341149413747189e-13 | 3.976082366916658e-16 | 3.3378845753551785e-16 |
| Head | 1.3946565841013234 | 1.5432100042289676e-13 | 2.2315482794965646e-12 | 7.183142969324763e-14 | 5.029310301551959e-13 | 1.6283882499911915e-15 | 1.0742966659288627e-15 |
| Whole body | 1.463136745675353 | 2.0061730054976579e-13 | 3.2640556923979602e-12 | 9.214851104388799e-14 | 7.37632177560954e-13 | 2.4486080463777027e-15 | 1.4985537998499074e-15 |
| Go:0022403~cell cycle phase | 1.928899643580963 | 3.297362383136715e-13 | 4.7628567756419216e-12 | 2.6677549058717887e-12 | 1.0671130645789617e-11 | 2.597464140378801e-15 | 9.65726934942202e-16 |
| Cell division | 2.043120298891528 | 2.0383694732117874e-13 | 3.963496197911809e-12 | 3.3573144264664734e-13 | 1.6786572132332367e-12 | 2.6743742804732204e-15 | 9.659346320160195e-16 |
| Go:0000279~m phase | 1.9948565816517663 | 6.961098364399732e-13 | 1.078026556911027e-11 | 4.825251309625855e-12 | 2.4126034503524352e-11 | 5.772924668904538e-15 | 2.1491744369336008e-15 |
| Go:0000278~mitotic cell cycle | 2.0743576494903473 | 2.7478019859472624e-12 | 1.4721557306529576e-11 | 5.490274901376324e-12 | 3.294131634135056e-11 | 7.91346417031652e-15 | 2.728394311538057e-15 |
| Thymus | 1.297653121996136 | 2.1296298058359753e-12 | 2.969846590872294e-11 | 7.450706718259426e-13 | 6.7057470687359455e-12 | 2.2160484024469804e-14 | 1.5295940048257806e-14 |
| Ribonucleoprotein | 1.984025781000403 | 1.766586876783549e-12 | 3.4994229736184934e-11 | 2.4713564528155985e-12 | 1.482813871689359e-11 | 2.349589182974304e-14 | 8.680825465087358e-15 |
| Nucleus | 1.2126628165121947 | 5.005329484220056e-12 | 4.523048602322888e-11 | 2.737809978725636e-12 | 1.9164669851079452e-11 | 3.038631995425044e-14 | 2.4569769701697746e-14 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.3462867209051432 | 9.578907807672228e-43 | 9.867462218087801e-42 | 3.8525448030728734e-42 | 3.8525448030728734e-42 | 6.723463879708331e-45 | 4.695543042976582e-45 |
| Brain | 1.3165980848607381 | 7.69817865110467e-39 | 1.005850343065025e-37 | 2.033185239602093e-38 | 2.033185239602093e-38 | 7.672397130573935e-41 | 5.53825802237746e-41 |
| Acetylation | 1.5807420294116445 | 2.1165794975111852e-30 | 2.6744969604266977e-29 | 5.221007761670859e-30 | 1.0442015523341719e-29 | 1.822341278070108e-32 | 1.037538969368228e-32 |
| Cytoplasm | 1.390199084504932 | 1.7045564258874579e-19 | 1.8275500081530276e-18 | 2.378429269402072e-19 | 7.135287808206215e-19 | 1.2452509263885192e-21 | 8.355668754350284e-22 |
| Embryo | 1.5125822701396314 | 8.915330355686825e-19 | 1.3824759074224274e-17 | 1.3972404684536044e-18 | 2.7944809369072088e-18 | 1.0545211082668713e-20 | 6.413906730709946e-21 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.4507903229026546 | 3.2125986783028352e-15 | 2.105590906601227e-14 | 8.31972423847334e-15 | 8.31972423847334e-15 | 1.4319663061055662e-17 | 9.126700790633054e-18 |
| Go:0043228~non-membrane-bounded organelle | 1.4507903229026546 | 3.2125986783028352e-15 | 2.105590906601227e-14 | 8.31972423847334e-15 | 8.31972423847334e-15 | 1.4319663061055662e-17 | 9.126700790633054e-18 |
| Bone marrow | 1.547887856448082 | 9.259260025373806e-14 | 1.5987211554602254e-12 | 1.0791367799356522e-13 | 3.2363001167823313e-13 | 1.1944903161392569e-15 | 6.452924697549102e-16 |
| Go:0051301~cell division | 2.3180907656748726 | 4.39648317751562e-13 | 3.2751579226442118e-12 | 6.5920602310143295e-12 | 6.5920602310143295e-12 | 1.779409437894093e-15 | 6.364057780097211e-16 |
| Mmu04360:axon guidance | 3.096508958182068 | 4.929390229335695e-14 | 2.5979218776228663e-12 | 3.7969627442180354e-13 | 3.7969627442180354e-13 | 2.102739473048343e-15 | 4.751831302545656e-16 |
| Mammary tumor | 1.3724194029832 | 2.9320990080350384e-13 | 4.5075054799781356e-12 | 2.2803980925800715e-13 | 9.12048214729566e-13 | 3.3968030387031006e-15 | 2.1395965763349247e-15 |
| Cerebellum | 1.505978044052698 | 3.240741008880832e-13 | 4.951594689828198e-12 | 2.000621890374532e-13 | 1.000310945187266e-12 | 3.801681965132337e-15 | 2.2995089821393604e-15 |
| Cell cycle | 1.9417222976997344 | 3.4425795547576854e-12 | 5.767608612927688e-11 | 5.629940957874169e-12 | 2.25199858761016e-11 | 3.934894852141963e-14 | 1.6919762382463323e-14 |
| Go:0005578~proteinaceous extracellular matrix | 2.174011071290361 | 8.08952904662874e-12 | 1.836530927334934e-10 | 3.6283420712379666e-11 | 7.25669524470618e-11 | 1.2489609247016723e-13 | 4.594966729641003e-14 |
| Go:0031012~extracellular matrix | 2.1345207355137115 | 1.4948042803553108e-11 | 3.307465412660804e-10 | 4.3561709794914805e-11 | 1.3068501836244195e-10 | 2.249173369940255e-13 | 8.493492272803772e-14 |
| Mammary gland | 1.3942631381593693 | 2.3703705664956942e-11 | 3.2850389075633757e-10 | 1.106714719867341e-11 | 6.640299421434293e-11 | 2.50534684807452e-13 | 1.705819332152303e-13 |
| Cell division | 2.2249691416635016 | 4.1945114048758114e-11 | 8.54938342342848e-10 | 6.675837660452544e-11 | 3.3379199404492965e-10 | 5.825124149080101e-13 | 2.0564306486468046e-13 |
| Mitosis | 2.5002047497703193 | 3.750599830709689e-11 | 9.119593968875961e-10 | 5.934297497844909e-11 | 3.5605751680378717e-10 | 6.213569040096594e-13 | 1.8385640834598132e-13 |
| Go:0007049~cell cycle | 1.753896772234127 | 1.3867240689080518e-10 | 1.5534906694369965e-09 | 1.5637602324147792e-09 | 3.127520575851861e-09 | 8.42813286936603e-13 | 4.2018044943188185e-13 |
| Go:0000087~m phase of mitotic cell cycle | 2.4189499415453293 | 2.64301913688314e-10 | 4.685640764279242e-09 | 3.144412730193835e-09 | 9.433238190581505e-09 | 2.5419371259771236e-12 | 8.008806334980807e-13 |
| Go:0000280~nuclear division | 2.433011394465545 | 5.773315159274262e-10 | 5.162681393500179e-09 | 2.5984047136162758e-09 | 1.0393618965487406e-08 | 2.800712441491591e-12 | 8.747383799531339e-13 |
| Go:0007067~mitosis | 2.433011394465545 | 5.773315159274262e-10 | 5.162681393500179e-09 | 2.5984047136162758e-09 | 1.0393618965487406e-08 | 2.800712441491591e-12 | 8.747383799531339e-13 |
| Go:0000278~mitotic cell cycle | 2.2103177285786715 | 3.1101821118539874e-09 | 1.609503641475385e-08 | 6.480571967948379e-09 | 3.2402859506674986e-08 | 8.731612318936968e-12 | 3.141584041157408e-12 |
| Neurogenesis | 2.5899111701903195 | 1.492448831186266e-09 | 1.9171297882536464e-08 | 1.0692903229525541e-09 | 7.485032593734786e-09 | 1.3062862693847248e-11 | 3.657974042110316e-12 |
| Head | 1.4182127306117418 | 1.4456791319616968e-09 | 2.0162915781440915e-08 | 5.822355930718004e-10 | 4.075649262524905e-09 | 1.5379804995142624e-11 | 1.0400614583320923e-11 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Brain | 1.3592246245550172 | 9.575795940662816e-29 | 1.2390761328666498e-27 | 2.2066665163294913e-28 | 2.2066665163294913e-28 | 9.67836191372584e-31 | 6.889061827814975e-31 |
| Phosphoprotein | 1.3548450006349937 | 1.23233769579151e-25 | 1.223859032512793e-24 | 4.300473418437235e-25 | 4.300473418437235e-25 | 8.48219609159218e-28 | 6.04087105780152e-28 |
| Acetylation | 1.5386861976039916 | 6.716542058650603e-15 | 1.5543122344752192e-13 | 2.8199664825478976e-14 | 5.6288307348495437e-14 | 1.6452040158365282e-16 | 3.292422577769903e-17 |
| Mmu04360:axon guidance | 3.542135296686089 | 1.390088044672666e-11 | 7.167821891584936e-10 | 9.988010418737758e-11 | 9.988010418737758e-11 | 5.874822509698364e-13 | 1.2525498978855384e-13 |
| Cerebellum | 1.6203312734039375 | 4.770062123071739e-11 | 7.662537271357905e-10 | 6.82314205135981e-11 | 1.364628410271962e-10 | 5.985636426536857e-13 | 3.4321021666568584e-13 |
| Head | 1.588557371411172 | 1.5819445753351147e-10 | 2.417177569213891e-09 | 1.4349144095149313e-10 | 4.304743228544794e-10 | 1.8879918761560227e-12 | 1.1380529738686888e-12 |
| Cytoplasm | 1.3675487655721736 | 2.041676161468331e-09 | 2.0661206079353178e-08 | 2.4200219606029805e-09 | 7.260065881808941e-09 | 1.4319657150491277e-11 | 1.0008249244255186e-11 |
| Go:0051301~cell division | 2.466311191516205 | 1.4381262736229417e-08 | 1.1358175511233526e-07 | 1.9225633640651552e-07 | 1.9225633640651552e-07 | 6.295230399614817e-11 | 2.178976287678082e-11 |
| Cell cycle | 2.0565584736400933 | 3.013964766651611e-08 | 4.981543844095881e-07 | 4.37611606951549e-08 | 1.7504463112327784e-07 | 3.4525574928109153e-10 | 1.4774335236898637e-10 |
| Embryo | 1.4551281769590545 | 3.609369059187628e-08 | 5.119324852920215e-07 | 2.279247102165982e-08 | 9.116988086699251e-08 | 3.9986795737783833e-10 | 2.5966681438456395e-10 |
| Go:0005578~proteinaceous extracellular matrix | 2.317835393647668 | 5.222308674390774e-08 | 1.1499402319259389e-06 | 3.882722883208345e-07 | 3.882722883208345e-07 | 8.022156197091118e-10 | 2.967220500545198e-10 |
| Go:0007156~homophilic cell adhesion | 3.301546465426064 | 2.8086020242845677e-07 | 1.6291160087611445e-06 | 1.3787764789885415e-06 | 2.757551056942198e-06 | 9.02932153213853e-10 | 2.1277295859159523e-10 |
| Cell division | 2.422805521091812 | 6.489445703028451e-08 | 1.3033503609705122e-06 | 9.159589819951464e-08 | 4.579794072867571e-07 | 9.033125993611611e-10 | 3.1811012150890727e-10 |
| Go:0031012~extracellular matrix | 2.2643440468262592 | 1.0039192976929456e-07 | 2.1481368128739575e-06 | 3.6265450353845097e-07 | 7.253088756264958e-07 | 1.4985725541187987e-09 | 5.704087514818704e-10 |
| Cell adhesion | 2.098550346088546 | 2.0240538001203845e-07 | 3.437829698693662e-06 | 2.013343806117618e-07 | 1.2080056753793755e-06 | 2.3826556295895905e-09 | 9.921833443724219e-10 |
| Go:0007155~cell adhesion | 1.8834081519503516 | 3.976163942542854e-07 | 4.5733173248407866e-06 | 2.580368183235926e-06 | 7.741084574908186e-06 | 2.534746130373928e-09 | 1.2048983741344673e-09 |
| Go:0022610~biological adhesion | 1.8800568918935003 | 4.3827703222198977e-07 | 5.000295899559859e-06 | 2.1159597981545986e-06 | 8.463812329106979e-06 | 2.771397507028847e-09 | 1.3281124540899664e-09 |
| Cholesterol biosynthesis | 7.99910394265233 | 1.4354072241928861e-07 | 4.479912996124824e-06 | 2.2488286777200273e-07 | 1.5741790125867183e-06 | 3.10489203699119e-09 | 1.4072626047932224e-10 |
| Go:0005886~plasma membrane | 1.3164770510906105 | 3.969765403066816e-07 | 4.456610280811901e-06 | 5.015849667833194e-07 | 1.5047541457313685e-06 | 3.108998456402646e-09 | 2.2555489375838182e-09 |
| Neurogenesis | 2.922749517507582 | 3.783745723406895e-07 | 4.8489469994805745e-06 | 2.1298169794459199e-07 | 1.7038523130175065e-06 | 3.3606583276141076e-09 | 9.273888762868672e-10 |
| Cytoskeleton | 1.842780773699824 | 4.2950530609875415e-07 | 6.242048122828692e-06 | 2.4370780271620873e-07 | 2.1933680858232663e-06 | 4.326174555223146e-09 | 2.1054185637757368e-09 |
| Go:0043228~non-membrane-bounded organelle | 1.4113849158048997 | 1.0996377667327195e-06 | 6.584289646838215e-06 | 5.557891153973316e-07 | 2.2231546082940312e-06 | 4.593299760607611e-09 | 3.123972597983784e-09 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.4113849158048997 | 1.0996377667327195e-06 | 6.584289646838215e-06 | 5.557891153973316e-07 | 2.2231546082940312e-06 | 4.593299760607611e-09 | 3.123972597983784e-09 |
| Go:0007049~cell cycle | 1.8222553082013697 | 8.557457755742348e-07 | 9.282784352304674e-06 | 3.142532238764595e-06 | 1.571256243926289e-05 | 5.1449527603824945e-09 | 2.593170076459999e-09 |
| Go:0005874~microtubule | 2.3981046931407946 | 4.537180753327874e-07 | 1.033180733189809e-05 | 6.976978319528371e-07 | 3.4884842918803116e-06 | 7.207624734331084e-09 | 2.577944169505657e-09 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Lens | 36.90650154798762 | 0.04728340373663664 | 2.9083321306725196 | 0.1734718700392065 | 0.1734718700392065 | 0.002882524302608968 | 6.969224790095509e-05 |
| Eye lens protein | 35.708 | 0.075630898915506 | 3.547372289881734 | 0.3435926036381868 | 0.3435926036381868 | 0.003068084158623029 | 7.709881896385974e-05 |
| Go:0005576~extracellular region | 2.2328571428571427 | 0.26638563689030603 | 4.217497007231142 | 0.2722108098386411 | 0.2722108098386411 | 0.004065353047557482 | 0.0017585189183363472 |
| Methylation | 5.385822021116139 | 0.6207469955219229 | 14.651299213298364 | 0.6027766399494308 | 0.8422136022301359 | 0.013387773418642246 | 0.0023735310106655304 |
| Secreted | 2.011718309859155 | 0.9524500740316248 | 31.32036505808886 | 0.7676981855881923 | 0.9874640339958141 | 0.03145915541027876 | 0.014820331849500146 |
| Eye lens | 59.51333333333334 | 0.9757393030818927 | 32.33690930721627 | 0.6796174437840115 | 0.989464007602989 | 0.03268711213448799 | 0.00047961890489144407 |
| Mmu05322:systemic lupus erythematosus | 9.284789644012944 | 0.3423123655025787 | 26.23274108066903 | 0.6188720083230714 | 0.6188720083230714 | 0.03642096955469927 | 0.003767885260400599 |
| Testicle | 4.999811261402956 | 0.9070866074821837 | 37.486625441513596 | 0.780473289950075 | 0.9518080235746562 | 0.04490829816002522 | 0.008510654854181565 |
| Oxidation | 36.62358974358975 | 0.9823280550544471 | 46.9991269559106 | 0.7723438948594297 | 0.9993884999354655 | 0.05257892719577836 | 0.0013180120073125784 |
| Citrullination | 29.75666666666667 | 0.9149254305134036 | 54.22509623949641 | 0.780848853365526 | 0.9998892197135542 | 0.06431974840060664 | 0.0020112322067284594 |
| Sodium | 6.933592233009709 | 0.9995209415325651 | 56.35754456493463 | 0.7485648485405259 | 0.9999364695467566 | 0.06810961294303017 | 0.00932352843764296 |
| Go:0006814~sodium ion transport | 6.794 | 0.999999900764064 | 62.81424556193909 | 0.9999999999954428 | 0.9999999999954428 | 0.06941347007352272 | 0.009725588858666673 |
| Go:0043010~camera-type eye development | 6.532692307692308 | 0.999999999987718 | 65.41088156020788 | 0.999999178872848 | 0.9999999999993258 | 0.07429932418702033 | 0.01081678479629359 |
| Symport | 6.551926605504587 | 0.9999856220958191 | 60.0706756939286 | 0.7375115796619623 | 0.999977463773737 | 0.07513269966358997 | 0.0108716656810391 |
| Sodium transport | 6.433873873873874 | 0.9907733979888411 | 61.267189731117355 | 0.7072195714208029 | 0.9999841919471342 | 0.07752348878971994 | 0.011418776863142412 |
| Go:0044421~extracellular region part | 2.2617054263565897 | 0.9999930665246635 | 58.781435479624264 | 0.961905637444142 | 0.9985488195414628 | 0.08037278921299093 | 0.03318448593563117 |
| Thiol protease inhibitor | 21.64121212121212 | 0.9957023591875529 | 65.85838319687187 | 0.7142265184243817 | 0.9999963673859482 | 0.08737245776561037 | 0.003809036010216497 |
| Intestinal mucosa | 21.249197860962568 | 0.4233285274504459 | 61.462249295920635 | 0.8714876751408596 | 0.9978775652829738 | 0.08904381258281606 | 0.003952474418290688 |
| Dna binding | 3.690749354005168 | 0.9918156230162827 | 67.7409961504396 | 0.6984401186076856 | 0.9999981245339707 | 0.09176574574164964 | 0.023281221976960302 |
| Go:0009952~anterior/posterior pattern formation | 5.550653594771242 | 0.9961766586166241 | 75.82029426201508 | 0.9999962439305908 | 1.0 | 0.09809023788723005 | 0.016727099542376892 |
| Mmu04062:chemokine signaling pathway | 5.254578754578755 | 0.9823871659609076 | 57.734221384658134 | 0.7446641348085712 | 0.9348035959469445 | 0.09968763047069695 | 0.01802975130314745 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Signal | 1.801264145668478 | 4.31528619837529e-06 | 5.216494187720855e-05 | 1.184319755243024e-05 | 1.184319755243024e-05 | 3.8958116585206164e-08 | 2.1153409129792148e-08 |
| Glycoprotein | 1.5755635780184516 | 0.0006094192663751441 | 0.006438354577853467 | 0.0007306222260460293 | 0.0014607106432548145 | 4.808470416548162e-06 | 2.988255524214441e-06 |
| Collagen | 7.673199243596355 | 0.00014086409008806644 | 0.007923957834943263 | 0.0005995157499776305 | 0.001797469208007052 | 5.918030495670055e-06 | 6.90558645749673e-07 |
| Go:0007155~cell adhesion | 2.742003834123701 | 0.002398410918052263 | 0.034148933255440106 | 0.02725528459942894 | 0.02725528459942894 | 2.0871081116744422e-05 | 7.2766150788458376e-06 |
| Go:0022610~biological adhesion | 2.7371248237426977 | 0.0024712412063108147 | 0.03512342039246885 | 0.014110648924832603 | 0.028022187436585355 | 2.146676457296386e-05 | 7.497849960184852e-06 |
| Secreted | 1.9518025118218338 | 0.0035279900979144907 | 0.04677355604163491 | 0.0026519064399834757 | 0.010565504663148872 | 3.493924451645969e-05 | 1.732449763075019e-05 |
| Mmu04512:ecm-receptor interaction | 6.6902448503692185 | 0.0014204803550132983 | 0.05173467962440448 | 0.004348318697234621 | 0.004348318697234621 | 4.736627090554126e-05 | 6.403088480572384e-06 |
| Go:0044421~extracellular region part | 2.296549627602979 | 0.007903006234099896 | 0.06839664961590941 | 0.011576783136078461 | 0.011576783136078461 | 5.390741640260289e-05 | 2.2540656476447365e-05 |
| Mmu04510:focal adhesion | 4.0509395025524055 | 0.001679666795020407 | 0.07151670422143042 | 0.0030078214914339707 | 0.0060065959927436685 | 6.548381404980217e-05 | 1.5144741325191191e-05 |
| Spleen | 2.1346226161697555 | 0.00638970554862428 | 0.11666599392881061 | 0.012457032906786236 | 0.012457032906786236 | 0.00010108579510741648 | 4.611553446150152e-05 |
| Disulfide bond | 1.618550897555683 | 0.013680713508096853 | 0.1502639303083808 | 0.00680490504118525 | 0.03356459830193781 | 0.00011229902267930086 | 6.752299878033317e-05 |
| Go:0005576~extracellular region | 1.7147759103641453 | 0.02044658428177204 | 0.26323059734982834 | 0.02217930322519568 | 0.04386668495883628 | 0.0002076540145738177 | 0.00011737100595171302 |
| Hydroxylation | 7.049751805054152 | 0.023605160116721646 | 0.610391199154714 | 0.022901161484123023 | 0.12977614899884415 | 0.0004571481089340915 | 5.854785786076796e-05 |
| Extracellular matrix | 3.6312604871103877 | 0.05110885492559414 | 0.6623761298699571 | 0.021324041634688706 | 0.14005150927056198 | 0.0004962018051875424 | 0.00012857308628561726 |
| Go:0005886~plasma membrane | 1.4553661795069026 | 0.05940013164672242 | 0.6570495150025946 | 0.03670648008404154 | 0.1061268002631488 | 0.0005192695246233239 | 0.0003478795294677561 |
| Cell adhesion | 2.7138894166825005 | 0.06103952749135877 | 1.180291656226451 | 0.03313381553410377 | 0.2362856155044395 | 0.0008863221224474393 | 0.0003086871304623123 |
| Go:0044459~plasma membrane part | 1.6139908504736862 | 0.1452416263691989 | 1.8736865096137922 | 0.0773232814256678 | 0.27523326900630185 | 0.001489192949412662 | 0.0008912869649113538 |
| Go:0042330~taxis | 4.70417171542323 | 0.32991782389676816 | 2.4927021052117193 | 0.4937767478315044 | 0.8702742265527903 | 0.0015413580846178427 | 0.00030325319370804756 |
| Go:0006935~chemotaxis | 4.70417171542323 | 0.32991782389676816 | 2.4927021052117193 | 0.4937767478315044 | 0.8702742265527903 | 0.0015413580846178427 | 0.00030325319370804756 |
| Transmembrane protein | 2.6130354180895696 | 0.13296926106801754 | 2.5579032073026253 | 0.06327500796080832 | 0.44472391785869936 | 0.0019332927367402095 | 0.0006991713895589109 |
| Inflammation | 15.165852622637503 | 0.11074769801174056 | 2.7967895589606995 | 0.06237176274304457 | 0.4748216925662103 | 0.0021162360321837098 | 0.00011506620238824683 |
| Go:0048584~positive regulation of response to stimulus | 3.4459322377764257 | 0.5932037465306256 | 4.01577628907821 | 0.5635234615965337 | 0.9637052660337884 | 0.0025014602434358896 | 0.0006811639816620054 |
| Chemotaxis | 6.237568417375102 | 0.21336659297116856 | 3.536889632396334 | 0.07162729737415652 | 0.5584844698853126 | 0.002685670287688925 | 0.0003920684637525758 |
| Go:0009611~response to wounding | 2.585938774400522 | 0.3071353894139851 | 4.8596518267844395 | 0.5534078366364628 | 0.9822353976789047 | 0.003039591177587312 | 0.0011112628956284729 |
| Go:0005578~proteinaceous extracellular matrix | 2.682907506436918 | 0.1873285208023575 | 4.188561599182106 | 0.1355320154264802 | 0.5172252257692818 | 0.003365643112629854 | 0.0011778758483899933 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| None |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| None |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.3511017800799008 | 1.4333611888338436e-84 | 1.602820955564375e-83 | 6.959098295811154e-84 | 6.959098295811154e-84 | 1.0739349221930794e-86 | 7.026280337420802e-87 |
| Brain | 1.2888174095839213 | 9.819442962441144e-62 | 1.34753768314367e-60 | 3.237828753673533e-61 | 3.237828753673533e-61 | 9.962550011303177e-64 | 7.064347454993629e-64 |
| Acetylation | 1.5552031238266877 | 1.0478679326822636e-54 | 1.4596779672149264e-53 | 3.1688013620029386e-54 | 6.337602724005877e-54 | 9.78025111729302e-57 | 5.136607513148351e-57 |
| Embryo | 1.4848996917179411 | 1.1614831111530924e-32 | 1.9610705370823034e-31 | 2.356004827313651e-32 | 4.712009654627302e-32 | 1.4498491245007083e-34 | 8.355993605417931e-35 |
| Cytoplasm | 1.3305121157042337 | 2.0268006297908803e-27 | 2.2142445552694473e-26 | 3.2045886467834808e-27 | 9.613765940350442e-27 | 1.4836058549923522e-29 | 9.935297204857257e-30 |
| Nucleus | 1.279079648304467 | 2.920215765984635e-26 | 3.009779790583202e-25 | 3.2669515173700845e-26 | 1.3067806069480338e-25 | 2.016636739117336e-28 | 1.4314783166591348e-28 |
| Mammary tumor | 1.3619458101291437 | 7.835332477540374e-25 | 1.1636685760379768e-23 | 9.320109367807044e-25 | 2.7960328103421137e-24 | 8.603177877975734e-27 | 5.636929839957104e-27 |
| Head | 1.4725444354841266 | 2.4168190617652844e-24 | 4.028558691840047e-23 | 2.41992920336582e-24 | 9.67971681346328e-24 | 2.9783744041425475e-26 | 1.7387187494714278e-26 |
| Go:0043228~non-membrane-bounded organelle | 1.3951468171104446 | 7.863587431539927e-22 | 5.282225643573573e-21 | 2.2873053879050755e-21 | 2.2873053879050755e-21 | 3.540720414713739e-24 | 2.2339737021420247e-24 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.3951468171104446 | 7.863587431539927e-22 | 5.282225643573573e-21 | 2.2873053879050755e-21 | 2.2873053879050755e-21 | 3.540720414713739e-24 | 2.2339737021420247e-24 |
| Bone marrow | 1.4854547446072959 | 1.0367552646745411e-21 | 1.7508905721624527e-20 | 8.413989322893129e-22 | 4.206994661446565e-21 | 1.2944598958297122e-23 | 7.45867096888159e-24 |
| Whole body | 1.50488088378329 | 4.0352518819107823e-20 | 6.949902674034005e-19 | 2.7831744515329097e-20 | 1.6699046709197458e-19 | 5.138168218214603e-22 | 2.903058907849484e-22 |
| Go:0007049~cell cycle | 1.7122442995918519 | 7.830481936915233e-20 | 9.872614429682476e-19 | 2.3409835694867324e-18 | 2.3409835694867324e-18 | 5.25119688085853e-22 | 2.372873314216737e-22 |
| Go:0051301~cell division | 2.0227676023900267 | 3.386976942833366e-17 | 2.883977268239074e-16 | 3.419227727268814e-16 | 6.838455454537628e-16 | 1.5339738570070948e-19 | 5.131783246717221e-20 |
| Cerebellum | 1.4066688465716375 | 2.4642098903332342e-17 | 3.835872318265586e-16 | 1.3166764242727407e-17 | 9.216734969909185e-17 | 2.8359184522797492e-19 | 1.772812870743334e-19 |
| Mammary gland | 1.3392180434761098 | 5.5141521754903315e-17 | 7.990853098748885e-16 | 2.4000269392421783e-17 | 1.9200215513937426e-16 | 5.907758619673054e-19 | 3.9670159535901665e-19 |
| Thymus | 1.3062112643022765 | 9.677099682361629e-17 | 1.3531069257588686e-15 | 3.612455984488794e-17 | 3.251210386039914e-16 | 1.0003724264738197e-18 | 6.961942217526351e-19 |
| Cell cycle | 1.7545973683534144 | 1.78585088926654e-16 | 3.0435053433861674e-15 | 2.642847009820013e-16 | 1.3214235049100064e-15 | 2.0392338038734667e-18 | 8.754171025816372e-19 |
| Go:0022402~cell cycle process | 1.81312094728374 | 5.59389367569968e-16 | 7.781458953873061e-15 | 6.150436876809515e-15 | 1.8451310630428546e-14 | 4.138921182240589e-18 | 1.6951192956665698e-18 |
| Spinal ganglion | 1.8388547739918863 | 5.723647797193859e-16 | 1.3605146245375058e-14 | 3.269009413409649e-16 | 3.269009413409649e-15 | 1.005849050279892e-17 | 4.117732228197021e-18 |
| Cell division | 1.9953272837350757 | 1.0635296379815745e-15 | 2.2973372252212795e-14 | 1.6624226922579714e-15 | 9.974536153547827e-15 | 1.5392802706092327e-17 | 5.213380578341052e-18 |
| Rna-binding | 1.7000531837067419 | 1.5909668412676964e-15 | 2.5663947216437495e-14 | 1.5918178572924782e-15 | 1.1142725001047347e-14 | 1.7195563273221216e-17 | 7.798857065037727e-18 |
| Mmu04360:axon guidance | 2.4723973890917135 | 3.6259265543958256e-15 | 1.3322676295501878e-13 | 2.0872192862952943e-14 | 2.0872192862952943e-14 | 1.4076823678330886e-16 | 3.266600499455699e-17 |
| Go:0022403~cell cycle phase | 1.8333789248774575 | 1.099120794378905e-13 | 2.0872192862952943e-12 | 1.237343560944737e-12 | 4.949374243778948e-12 | 1.0682206076085525e-15 | 3.6183019884629854e-16 |
| Go:0000278~mitotic cell cycle | 1.9750098142010246 | 4.39648317751562e-13 | 2.5091040356528538e-12 | 1.187827614046455e-12 | 5.9392490925347374e-12 | 1.3724881480188988e-15 | 4.366532580914204e-16 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.3846218138357627 | 1.2126176666805578e-75 | 1.3451349991472252e-74 | 5.503893729296093e-75 | 5.503893729296093e-75 | 9.09734500710098e-78 | 5.9442042484341075e-78 |
| Brain | 1.3348984830338857 | 5.08826277968881e-62 | 7.073884905024044e-61 | 1.6060190548124806e-61 | 1.6060190548124806e-61 | 5.2829574171463176e-64 | 3.6606207048120933e-64 |
| Acetylation | 1.5752081610146125 | 2.4599599428722742e-43 | 3.27428064664353e-42 | 6.698692967784584e-43 | 1.3397385935569167e-42 | 2.2144439562924245e-45 | 1.205862717094252e-45 |
| Embryo | 1.530001935183972 | 8.091813945835905e-29 | 1.3527697489279824e-27 | 1.5356300128725734e-28 | 3.071260025745147e-28 | 1.0102829032056403e-30 | 5.821448881896335e-31 |
| Cytoplasm | 1.366879068227571 | 5.291452696918148e-25 | 5.7650845590462735e-24 | 7.863005763612978e-25 | 2.3589017290838935e-24 | 3.8990111224527165e-27 | 2.5938493612343863e-27 |
| Head | 1.5049676478422418 | 1.4301311396113716e-20 | 2.3364659421973757e-19 | 1.7681980384990096e-20 | 5.304594115497029e-20 | 1.744932274834549e-22 | 1.0288713234614185e-22 |
| Go:0007049~cell cycle | 1.845485973744889 | 4.15010119028261e-20 | 5.412077060414377e-19 | 1.1942450570433686e-18 | 1.1942450570433686e-18 | 2.903586328819277e-22 | 1.2576064212977606e-22 |
| Nucleus | 1.2814578494119542 | 6.266322192633488e-20 | 6.255360225128013e-19 | 6.39877000066854e-20 | 2.559508000267416e-19 | 4.2305917359791995e-22 | 3.0717265650164153e-22 |
| Mammary tumor | 1.3759945719189215 | 5.63462103358266e-20 | 8.15330209217381e-19 | 4.627711185396044e-20 | 1.8510844741584176e-19 | 6.089093664994795e-22 | 4.0536841968220576e-22 |
| Go:0051301~cell division | 2.2640929737315885 | 1.3361245903745004e-18 | 1.2180635918747896e-17 | 1.3439077155800711e-17 | 2.6878154311601423e-17 | 6.53492689316835e-21 | 2.0244311975371218e-21 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.4174634644347677 | 3.0041775743410753e-18 | 1.9773726464519124e-17 | 8.056177198040337e-18 | 8.056177198040337e-18 | 1.3382354149568667e-20 | 8.534595381650782e-21 |
| Go:0043228~non-membrane-bounded organelle | 1.4174634644347677 | 3.0041775743410753e-18 | 1.9773726464519124e-17 | 8.056177198040337e-18 | 8.056177198040337e-18 | 1.3382354149568667e-20 | 8.534595381650782e-21 |
| Cerebellum | 1.4919701431307992 | 1.400062409353418e-18 | 2.2600680328465063e-17 | 1.0262288331398038e-18 | 5.131144165699019e-18 | 1.68787637029573e-20 | 1.0072391434197252e-20 |
| Spinal ganglion | 2.05836621598601 | 3.791471619690247e-18 | 9.819910368998033e-17 | 3.7157713149391165e-18 | 2.2294627889634702e-17 | 7.333759174221941e-20 | 2.7276774242375877e-20 |
| Cell cycle | 1.9245008992040484 | 9.71439403895253e-18 | 1.7521259360415882e-16 | 1.43383600284433e-17 | 7.169180014221649e-17 | 1.184988432102752e-19 | 4.761957862231633e-20 |
| Cell division | 2.2402393279797126 | 2.8192765352046525e-17 | 6.566587749463954e-16 | 4.4780884644862165e-17 | 2.68685307869173e-16 | 4.44107946891195e-19 | 1.3819983015709082e-19 |
| Go:0022402~cell cycle process | 1.9742137578929322 | 2.0748729872734775e-16 | 3.0063390860174432e-15 | 2.211292481114892e-15 | 6.633877443344676e-15 | 1.6129048002296805e-18 | 6.28749390082872e-19 |
| Thymus | 1.34558810411673 | 8.375117926286718e-16 | 1.1763013528603177e-14 | 3.815164433808885e-16 | 2.6706151036662197e-15 | 8.784918104165196e-18 | 6.025264695170301e-18 |
| Mitosis | 2.445095085404496 | 6.951723391447311e-16 | 1.9000369590622376e-14 | 1.1106268686832038e-15 | 7.774388080782425e-15 | 1.2850228232698225e-17 | 3.407707544827113e-18 |
| Go:0000278~mitotic cell cycle | 2.2258451016243677 | 7.519015075128667e-15 | 4.430431009153942e-14 | 2.444080282944387e-14 | 9.776321131777548e-14 | 2.3769319552097124e-17 | 7.59496472235219e-18 |
| Go:0022403~cell cycle phase | 2.018513188557847 | 1.0120854466186864e-14 | 2.1094237467877974e-13 | 9.137135492665038e-14 | 4.566347300283269e-13 | 8.149798613914189e-17 | 3.066925595814201e-17 |
| Go:0000279~m phase | 2.101874930223111 | 1.261376545065478e-14 | 4.107825191113079e-13 | 1.5221157667610896e-13 | 9.132694600566538e-13 | 2.1955776763943274e-16 | 3.822353166865085e-17 |
| Go:0000280~nuclear division | 2.3684330735479735 | 3.6637359812630166e-14 | 6.217248937900877e-13 | 1.957323192414151e-13 | 1.3699041900849807e-12 | 2.8871703513900605e-16 | 5.0904821175542057e-17 |
| Go:0007067~mitosis | 2.3684330735479735 | 3.6637359812630166e-14 | 6.217248937900877e-13 | 1.957323192414151e-13 | 1.3699041900849807e-12 | 2.8871703513900605e-16 | 5.0904821175542057e-17 |
| Go:0000087~m phase of mitotic cell cycle | 2.346261464019559 | 3.6637359812630166e-14 | 6.217248937900877e-13 | 1.7119639039719914e-13 | 1.3699041900849807e-12 | 3.4868872740394133e-16 | 6.92530992622425e-17 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Phosphoprotein | 1.4123629874327703 | 3.2557163016475736e-60 | 3.541221863345701e-59 | 1.3346341042658494e-59 | 1.3346341042658494e-59 | 2.426607462301544e-62 | 1.5959393635527321e-62 |
| Brain | 1.3654673464117413 | 2.199576467507712e-51 | 3.016733933488186e-50 | 6.233674242559205e-51 | 6.233674242559205e-51 | 2.291792000940884e-53 | 1.5824291133149007e-53 |
| Acetylation | 1.5913046322417106 | 2.0254665392698065e-31 | 2.5667445543927797e-30 | 4.836840152108868e-31 | 9.673680304217737e-31 | 1.7588509644032247e-33 | 9.928757545440228e-34 |
| Cytoplasm | 1.3783376661850526 | 2.4167535878830835e-18 | 2.5486802064985447e-17 | 3.201866150277049e-18 | 9.605598450831146e-18 | 1.746472445605663e-20 | 1.184683131315237e-20 |
| Embryo | 1.5010955247185636 | 5.9885755437425786e-18 | 9.233412988043294e-17 | 9.53980198179491e-18 | 1.907960396358982e-17 | 7.014560280731551e-20 | 4.308327729311208e-20 |
| Head | 1.5411066204790158 | 3.1395643333558004e-16 | 5.003004798801788e-15 | 3.446011794796657e-16 | 1.033803538438997e-15 | 3.800748303084548e-18 | 2.2586793765149643e-18 |
| Cerebellum | 1.5554864859781443 | 3.5131004268301788e-16 | 5.664211769124426e-15 | 2.926082626780145e-16 | 1.170433050712058e-15 | 4.30306268644139e-18 | 2.5274103790145173e-18 |
| Go:0007049~cell cycle | 1.8875654159342103 | 6.40640166292472e-15 | 7.889098025820994e-14 | 1.5500559199511154e-13 | 1.5500559199511154e-13 | 4.292594627391624e-17 | 1.941333837249915e-17 |
| Cell cycle | 2.0350592458383305 | 7.934375568629711e-15 | 3.219646771412954e-13 | 3.0531133177191805e-14 | 1.2212453270876722e-13 | 2.0710811993581918e-16 | 3.889399788543976e-17 |
| Go:0051301~cell division | 2.363069870978135 | 3.6637359812630166e-14 | 4.107825191113079e-13 | 4.0090153419214403e-13 | 8.018030683842881e-13 | 2.586632578789713e-16 | 4.8515623368110257e-17 |
| Mitosis | 2.6547571000700976 | 2.0383694732117874e-13 | 5.3512749786932545e-12 | 4.030109579389318e-13 | 2.015054789694659e-12 | 3.666730983055016e-15 | 9.461511068978676e-16 |
| Mmu04360:axon guidance | 3.104045200456813 | 9.85878045867139e-14 | 4.907185768843192e-12 | 7.074341112911497e-13 | 7.074341112911497e-13 | 3.9622582056613314e-15 | 9.264319263330562e-16 |
| Go:0022402~cell cycle process | 2.0631210798018826 | 8.426592756904938e-13 | 1.142419492339286e-11 | 7.483458297485868e-12 | 2.2450485914760065e-11 | 6.2367848720371925e-15 | 2.5616280017018378e-15 |
| Cell division | 2.3335788973384033 | 4.756195437494171e-13 | 1.0202949596305189e-11 | 6.411537967210279e-13 | 3.8469227803261674e-12 | 6.955880932295592e-15 | 2.3481060668360045e-15 |
| Spinal ganglion | 2.092666503658284 | 7.098766019453251e-13 | 1.680877659282487e-11 | 6.945555242054979e-13 | 3.4727776210274897e-12 | 1.279715628523308e-14 | 5.1427011884235675e-15 |
| Go:0000087~m phase of mitotic cell cycle | 2.5580904501293964 | 2.1249668691325496e-12 | 4.0401015866109447e-11 | 1.9844570431359898e-11 | 7.937850377004452e-11 | 2.202894333501581e-14 | 6.386331264322975e-15 |
| Go:0000280~nuclear division | 2.5751570283734027 | 4.39648317751562e-12 | 4.305444889496357e-11 | 1.6918022538447985e-11 | 8.459022371454239e-11 | 2.339909985587028e-14 | 6.638331163357255e-15 |
| Go:0007067~mitosis | 2.5751570283734027 | 4.39648317751562e-12 | 4.305444889496357e-11 | 1.6918022538447985e-11 | 8.459022371454239e-11 | 2.339909985587028e-14 | 6.638331163357255e-15 |
| Go:0000279~m phase | 2.247575215576786 | 3.4439118223872356e-12 | 5.467848396278896e-11 | 1.7906898186481612e-11 | 1.074416111634946e-10 | 2.97040777602861e-14 | 1.0381571709789114e-14 |
| Go:0000278~mitotic cell cycle | 2.3490015516427176 | 1.3958834088612093e-11 | 7.855938122247608e-11 | 2.204958438056792e-11 | 1.5434709066397545e-10 | 4.277710873668106e-14 | 1.4104565756805733e-14 |
| Go:0022403~cell cycle phase | 2.109699638663053 | 2.1029844532449715e-11 | 3.046340957268967e-10 | 7.481826269639669e-11 | 5.98545990548871e-10 | 1.6574896531293242e-13 | 6.37687455668553e-14 |
| Go:0048285~organelle fission | 2.4836539867560736 | 3.605116205562808e-11 | 3.3176794644873553e-10 | 7.24295068366132e-11 | 6.518658945964262e-10 | 1.8049200207571305e-13 | 5.4628417898267706e-14 |
| Go:0043228~non-membrane-bounded organelle | 1.395202750103568 | 5.721290108340327e-11 | 3.4877656318599293e-10 | 1.324773624133968e-10 | 1.324773624133968e-10 | 2.3873154859281176e-13 | 1.6249553028781412e-13 |
| Go:0043232~intracellular non-membrane-bounded organelle | 1.395202750103568 | 5.721290108340327e-11 | 3.4877656318599293e-10 | 1.324773624133968e-10 | 1.324773624133968e-10 | 2.3873154859281176e-13 | 1.6249553028781412e-13 |
| Mammary tumor | 1.3333027618877717 | 1.487191481075456e-10 | 1.8010370972376677e-09 | 6.202682811817795e-11 | 3.721609687090677e-10 | 1.368194761520956e-12 | 1.0699181837075073e-12 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0006974~response to dna damage stimulus | 3.1917942293387624 | 0.004960504054106751 | 0.08588605712025466 | 0.08031462105937659 | 0.08031462105937659 | 5.1174513791554585e-05 | 1.5069123409257893e-05 |
| Go:0005578~proteinaceous extracellular matrix | 2.8171679621493744 | 0.013286539929254437 | 0.29774151708442664 | 0.05631526964054123 | 0.05631526964054123 | 0.00022907708074129768 | 7.599481514159628e-05 |
| Go:0044421~extracellular region part | 1.9818449036050836 | 0.04408814374608572 | 0.344136971413167 | 0.0329503152379208 | 0.06481490720156335 | 0.0002648297843952718 | 0.000128087167594753 |
| Mmu05020:prion diseases | 9.836571428571428 | 0.008614503371916049 | 0.34440426249494616 | 0.03707619120685035 | 0.03707619120685035 | 0.00029744385644725604 | 2.5981111991208046e-05 |
| Go:0031012~extracellular matrix | 2.70776338109503 | 0.02195069509767944 | 0.47195163864058864 | 0.03018763750083797 | 0.08785654192543124 | 0.00036340431137827495 | 0.00012610111794638654 |
| Dna repair | 3.475725538350954 | 0.02069175674679935 | 0.5157633034494347 | 0.11367097784955271 | 0.11367097784955271 | 0.00038421610792657967 | 0.00010248901624712811 |
| Go:0033554~cellular response to stress | 2.519375533058924 | 0.09188535221580685 | 0.6504487792268976 | 0.27234988786834013 | 0.47052531431478284 | 0.00038859802331450076 | 0.00014602667687280948 |
| Go:0006281~dna repair | 3.2093666700408274 | 0.04114401763969988 | 0.7309839812320362 | 0.2120309826903206 | 0.5107538412777373 | 0.00043687857748427755 | 0.0001273082295068626 |
| Colon | 1.7978977248372334 | 0.048223600735870176 | 0.7858638703370557 | 0.08986778056008027 | 0.08986778056008027 | 0.0006676167180391117 | 0.0003555133836655596 |
| Dna damage | 3.0994872068799744 | 0.06297066567093512 | 1.4194062719232625 | 0.15363019150823465 | 0.2836581472736125 | 0.0010618491445936312 | 0.0003187760947369657 |
| Salivary gland | 2.0616467403564176 | 0.11874732481390571 | 2.3244673953551342 | 0.13094880975081724 | 0.24475002872647866 | 0.001988845837992922 | 0.0009090174505572844 |
| Secreted | 1.563511639786908 | 0.23952335257586654 | 2.895075562902294 | 0.20428711786209208 | 0.49618723457883085 | 0.0021809004659385928 | 0.0013413049131487322 |
| Extracellular matrix | 2.8230071273930286 | 0.26996847547526215 | 3.098168949784197 | 0.16773288806098396 | 0.5202102859151705 | 0.002336143922542189 | 0.0007709466910356433 |
| Mmu04640:hematopoietic cell lineage | 4.781666666666667 | 0.12430895694845345 | 3.550129393657808 | 0.17956672068526802 | 0.3268892341928751 | 0.003112040270697373 | 0.0005977579419266186 |
| Go:0005576~extracellular region | 1.4664365374402548 | 0.3438414718896483 | 4.630153413187676 | 0.20577229753467086 | 0.602094333127237 | 0.0036358261757679956 | 0.002391186890923905 |
| Mmu03410:base excision repair | 7.1725 | 0.12275608910494995 | 5.354217144271689 | 0.18198504693267292 | 0.45262655108263006 | 0.004733830705054087 | 0.0005897818916487432 |
| Go:0030098~lymphocyte differentiation | 3.5713253170379127 | 0.8369521462389954 | 11.255810895496987 | 0.9454639297168513 | 0.999991154235709 | 0.007086975821102619 | 0.0018303547124770686 |
| Go:0006259~dna metabolic process | 2.0549965749464003 | 0.9338449863501694 | 13.856623813571666 | 0.945348834026129 | 0.9999995124746981 | 0.008844471908793297 | 0.004106325589290192 |
| Signal | 1.2926170164512134 | 0.7609804712783752 | 11.84619128283636 | 0.4448179769809003 | 0.9472555248186981 | 0.009326605102712455 | 0.006991182574106756 |
| Go:0006310~dna recombination | 4.071310861423221 | 0.9997159901304549 | 23.353840306881267 | 0.9866918695629862 | 0.9999999999944448 | 0.015716434754311975 | 0.0035290403122610354 |
| Go:0043281~regulation of caspase activity | 5.0891385767790265 | 0.9999999687299879 | 24.168172957079438 | 0.9787417810352802 | 0.999999999998038 | 0.016342423499242108 | 0.0029049653747213527 |
| Go:0052548~regulation of endopeptidase activity | 5.0891385767790265 | 0.9999999687299879 | 24.168172957079438 | 0.9787417810352802 | 0.999999999998038 | 0.016342423499242108 | 0.0029049653747213527 |
| Mmu04350:tgf-beta signaling pathway | 3.9572413793103447 | 0.7277097598059963 | 17.787724914556634 | 0.41505269897724406 | 0.8829242453563358 | 0.01674742097059136 | 0.0038989460278991135 |
| Go:0052547~regulation of peptidase activity | 4.9893515458617905 | 0.9999995738953429 | 25.61170992447197 | 0.9727780106681929 | 0.9999999999996985 | 0.017467790501965903 | 0.003169982461425763 |
| Go:0002521~leukocyte differentiation | 2.86712032494593 | 0.9012027705385219 | 30.809317797174895 | 0.9814543389251764 | 0.9999999999999998 | 0.021697382083526374 | 0.006989657060680294 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0032269~negative regulation of cellular protein metabolic process | 15.726851851851851 | 0.8313152040492973 | 18.45030892281162 | 0.9954687524829364 | 0.9954687524829364 | 0.014716890412909397 | 0.000898446366401668 |
| Go:0051248~negative regulation of protein metabolic process | 14.517094017094015 | 0.9272102207150612 | 21.14998679127075 | 0.9568801111973069 | 0.9981406751896433 | 0.01712513562134309 | 0.0011336338896280275 |
| Go:0005578~proteinaceous extracellular matrix | 4.784205693296602 | 0.4809435044360765 | 18.335636707447033 | 0.8011917779918656 | 0.8011917779918656 | 0.01860857316895191 | 0.0037188777088870903 |
| Protein synthesis inhibitor | 100.87005649717513 | 0.4954468276149132 | 20.363594625173608 | 0.9225321748438786 | 0.9225321748438786 | 0.019336501989925307 | 0.00015967026921150278 |
| Go:0031012~extracellular matrix | 4.598411297440424 | 0.5398354192796944 | 20.618437396469947 | 0.6017850581806332 | 0.8414248601117983 | 0.021185468715350042 | 0.004400352845683818 |
| Go:0001558~regulation of cell growth | 12.307971014492752 | 0.9852747813809432 | 27.74142310256631 | 0.943062863836185 | 0.9998154190582913 | 0.023342540223558462 | 0.0018243916506567586 |
| Go:0040008~regulation of growth | 5.897569444444444 | 0.7753072276475881 | 32.11320635697656 | 0.9228649508544438 | 0.9999645996918665 | 0.027763330067994498 | 0.00451408760807454 |
| Go:0015671~oxygen transport | 68.62626262626262 | 0.9999985224570703 | 32.32283792502352 | 0.8733242846869153 | 0.9999673812866126 | 0.02798188345509996 | 0.00036976991115488525 |
| Go:0015669~gas transport | 58.06837606837606 | 0.9999998480080233 | 36.96254664823206 | 0.8693155987819915 | 0.9999950187021118 | 0.032987117764075924 | 0.0005226551797770597 |
| Oxygen transport | 55.02003081664098 | 0.8941643374590672 | 34.13127594115798 | 0.9041589196659658 | 0.9908144873204052 | 0.03516917304733202 | 0.0005792614051161096 |
| Go:0044421~extracellular region part | 2.5701198026779424 | 0.9979903239846721 | 40.91357246172704 | 0.7531028065238476 | 0.9849495855389635 | 0.04762342890084129 | 0.017486726537079604 |
| Go:0043005~neuron projection | 4.639703153988869 | 0.8458460152907407 | 43.562846371704886 | 0.680357199058228 | 0.9895609804612672 | 0.051666353508513015 | 0.01056764812748014 |
| Go:0017148~negative regulation of translation | 34.31313131313131 | 0.9999999999984507 | 54.21252704696127 | 0.9478068827116163 | 0.9999999989449023 | 0.055202542096402636 | 0.0015248292239213104 |
| Heme | 6.393172594891383 | 0.9991946612705128 | 61.11941645407366 | 0.9709108001527463 | 0.9999753852558504 | 0.07781450927691248 | 0.011573433983450988 |
| Brain | 1.2893315131864083 | 0.9998300499230774 | 57.80922653340672 | 0.9956454812964031 | 0.9956454812964031 | 0.08143807427406735 | 0.06053628746186209 |
| Mammary gland | 1.7439752306170002 | 0.9986008927792317 | 59.9221848692578 | 0.9438713240454191 | 0.9968495717355856 | 0.08607191178539378 | 0.04617971597039522 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Secreted | 6.2866197183098596 | 0.8196137640839811 | 37.793228681088905 | 0.8196711591695311 | 0.8196711591695311 | 0.05375825676397095 | 0.00836022482526393 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0005576~extracellular region | 2.272628135223555 | 4.613231215355729e-05 | 0.0007491472880261085 | 0.00010693263922278984 | 0.00010693263922278984 | 6.110761385768216e-07 | 2.6212142183837594e-07 |
| Secreted | 2.2923585713174073 | 0.0011425226466110772 | 0.01700013258253419 | 0.003078761546766451 | 0.003078761546766451 | 1.3233863861881542e-05 | 5.6037873534967135e-06 |
| Signal | 1.6938333612366763 | 0.013102562915549454 | 0.14408928516762565 | 0.012990764157564949 | 0.025812768361732408 | 0.00011223303275496116 | 6.46506489926991e-05 |
| Go:0044421~extracellular region part | 2.46641813125037 | 0.052849920403290396 | 0.4838551307657313 | 0.03402571764867124 | 0.06689368583583544 | 0.0003955568090401712 | 0.0001542429879000915 |
| Chemotaxis | 9.545891997861343 | 0.023201983008660276 | 0.5132967924004528 | 0.030632488065458152 | 0.08911116019563492 | 0.00040049664306396083 | 3.8357738661173316e-05 |
| Go:0007626~locomotory behavior | 4.339966143920279 | 0.06719704942673754 | 0.7642227415205483 | 0.4002919413864574 | 0.4002919413864574 | 0.0004817993650233051 | 0.00010539035552335303 |
| Go:0006935~chemotaxis | 6.661250787870298 | 0.1083230399145384 | 0.9800921127036233 | 0.2798010698577851 | 0.4813135010220091 | 0.0006185227042877471 | 8.685332224820509e-05 |
| Go:0042330~taxis | 6.661250787870298 | 0.1083230399145384 | 0.9800921127036233 | 0.2798010698577851 | 0.4813135010220091 | 0.0006185227042877471 | 8.685332224820509e-05 |
| Epidermis | 12.608599079803268 | 0.1073469135270757 | 4.220506306646121 | 0.3448843123795732 | 0.3448843123795732 | 0.003803051244223619 | 0.0002722824797366145 |
| Go:0007610~behavior | 2.817227405522571 | 0.45874084403154325 | 8.42538640376448 | 0.8584964290058488 | 0.9971666371215738 | 0.005513763809540279 | 0.0018584439307292493 |
| Go:0043603~cellular amide metabolic process | 10.638481111763554 | 0.9732735658511147 | 9.255463723178725 | 0.8017643901484318 | 0.9984557177317753 | 0.006082459503835431 | 0.0005225332963042378 |
| Go:0042981~regulation of apoptosis | 2.4383860414394762 | 0.5799460736290072 | 10.19352737151179 | 0.7614447922865647 | 0.9992274181366189 | 0.006731035721825809 | 0.002624949324605077 |
| Go:0043067~regulation of programmed cell death | 2.407906215921483 | 0.6196700977590777 | 11.165875577612205 | 0.7315856042060227 | 0.9996260316681608 | 0.007410052380711871 | 0.002925156517061401 |
| Go:0010941~regulation of cell death | 2.3950754545577806 | 0.636531345631203 | 11.602358064509188 | 0.690943995419977 | 0.9997306851501826 | 0.007717127711158054 | 0.0030621572112134146 |
| Go:0043062~extracellular structure organization | 4.176853322403812 | 0.6521581718569831 | 20.368842781201025 | 0.8500676239705667 | 0.9999997446338155 | 0.014205921960482 | 0.0031949078680521335 |
| Go:0030198~extracellular matrix organization | 5.134910437608646 | 0.9998304875134757 | 22.65750907212366 | 0.8508332952603767 | 0.9999999634365588 | 0.01601014854404449 | 0.002919159887843664 |
| Heparin-binding | 7.444595017200042 | 0.5637722700110526 | 19.06131327620617 | 0.6166761146524085 | 0.9784094589393711 | 0.01632662526639004 | 0.0020312451440637437 |
| Mmu04621:nod-like receptor signaling pathway | 7.119106699751861 | 0.22299681537180138 | 17.33988012730897 | 0.7959873360304341 | 0.7959873360304341 | 0.017316159234006954 | 0.002270489072043894 |
| Go:0005615~extracellular space | 2.2414962429602183 | 0.7270035599757957 | 19.407846144753627 | 0.6418033807109198 | 0.9540416479287803 | 0.0174461351538642 | 0.0073495438344343455 |
| Go:0009611~response to wounding | 2.6902787249488527 | 0.8811633406423393 | 25.56325863617642 | 0.8602151625241159 | 0.999999997151602 | 0.018374343944802883 | 0.006433775696421451 |
| Pancreas | 2.1362910763173786 | 0.9036351020332886 | 19.470207431148225 | 0.6542171659436467 | 0.8804342316719564 | 0.018952236874068395 | 0.00838056074125531 |
| Go:0009897~external side of plasma membrane | 3.243459571629734 | 0.6499215464543767 | 22.48641434812302 | 0.5970719267114137 | 0.9736421423292907 | 0.02056272273723593 | 0.005945877865966037 |
| Go:0006769~nicotinamide metabolic process | 12.965648854961833 | 0.9999999787443417 | 29.64878826593298 | 0.881258707096445 | 0.9999999999338364 | 0.02184909308455087 | 0.0015284102441223296 |
| Go:0046496~nicotinamide nucleotide metabolic process | 12.965648854961833 | 0.9999999787443417 | 29.64878826593298 | 0.881258707096445 | 0.9999999999338364 | 0.02184909308455087 | 0.0015284102441223296 |
| Go:0009820~alkaloid metabolic process | 12.965648854961833 | 0.9999999787443417 | 29.64878826593298 | 0.881258707096445 | 0.9999999999338364 | 0.02184909308455087 | 0.0015284102441223296 |
| Termname | Foldenrichment | Easebonferroni | Afdr | Benjamini | Bonferroni | Ease | Fisher |
| Go:0019216~regulation of lipid metabolic process | 26.03065134099617 | 0.997676827490974 | 60.286705273306836 | 0.9999998506020262 | 0.9999998506020262 | 0.07017815747468342 | 0.0026220203656335244 |
| Mammary gland | 2.2578738879148 | 0.9852782264788486 | 49.91488946175334 | 0.9590316401793086 | 0.9590316401793086 | 0.07160805815038906 | 0.029892511406938275 |
| Lim domain | 19.56602739726027 | 0.8509640681617705 | 62.696046737228016 | 0.9972705008593026 | 0.9972705008593026 | 0.09370820252587478 | 0.0046547404331007194 |
| Developmental protein | 3.5093857493857494 | 0.9946164869306923 | 62.802037758810144 | 0.9481984963188085 | 0.9973166042163675 | 0.0939654781260521 | 0.02528472199624191 |
| None |
| TF | P-Value |
| Ctcf_(sc-15914) | 3.46792918002e-23 |
| Rad21 | 4.94810295276e-20 |
| Gata-1 | 1.17734588643e-14 |
| Bhlhe40_(nb100-1800) | 2.9217054127e-14 |
| Smc3_(ab9263) | 1.1783240947e-13 |
| Ctcf | 5.94395127873e-12 |
| P300_(sc-584) | 8.98352215529e-11 |
| Maz_(ab85725) | 5.69152900787e-10 |
| Hnf4a | 1.55671894486e-09 |
| Pol2-4h8 | 6.67135804713e-09 |
| Ets1 | 1.31285232463e-08 |
| Max | 2.338484487e-08 |
| Pparg | 6.14024337257e-08 |
| Pol2 | 8.61073052747e-08 |
| Mafk_(ab50322) | 9.70152440032e-08 |
| Jund | 3.63364588938e-07 |
| Srebf2 | 1.20748826279e-06 |
| Myod_(sc-32758) | 1.23684498703e-06 |
| Gata1_(sc-265) | 1.56697657792e-06 |
| P300 | 3.70087703356e-06 |
| C-myc | 3.79979687535e-06 |
| Znf384_(hpa004051) | 5.76239706146e-06 |
| Chd2_(ab68301) | 7.78676318301e-06 |
| Pol2(phosphos2) | 9.08630240572e-06 |
| Sin3a_(nb600-1263) | 1.00400075675e-05 |
| TF | P-Value |
| Srf | 4.8520228714e-48 |
| Creb | 1.40660203716e-44 |
| Hnf4 | 8.79065080619e-41 |
| Nrf-1 | 4.34825467634e-19 |
| Creb, atf | 1.44719593446e-18 |
| Gabpa | 6.62302049624e-14 |
| Sf1 | 1.31207791903e-13 |
| Myogenin | 1.5225564896e-10 |
| Usf | 4.49738913799e-10 |
| Pur1 | 8.47468385862e-10 |
| Atf2 | 2.58243347357e-09 |
| Ets2 | 1.0598734294e-08 |
| Ap-2rep | 1.97471624947e-08 |
| Pparg | 3.99415192788e-08 |
| Err2 (esrrb) | 4.12242629114e-08 |
| Stra13 | 6.29537458812e-08 |
| Gabp | 2.11292216256e-07 |
| Atf3 | 2.38423623817e-07 |
| Clock:bmal | 3.39120327316e-07 |
| Elk-1 | 4.96347579231e-07 |
| Esrrb | 7.21366813663e-07 |
| Fli1 | 1.58532659919e-06 |
| Smm_as_activator | 3.86660911317e-06 |
| Ap-1 | 7.05606581769e-06 |
| T3r | 1.0646883122e-05 |
| TF | P-Value |
| Srf | 3.04311017743e-13 |
| Fxr/rxr-alpha | 0.00189929633743 |
| Stat4 | 0.00253159312556 |
| Sf1 | 0.00281608540835 |
| Hoxc9 | 0.00305820111229 |
| Stat1:stat1 | 0.00463635749024 |
| Rest | 0.0249264791077 |
| Fra1 | 0.028534268049 |
| Nf-y | 0.0400136946424 |
| Hnf4 | 0.0400903766865 |
| Myb | 0.0591210952359 |
| Alpha-cp1 | 0.0708724103187 |
| Esrrb | 0.07449618165 |
| Hnf4, coup | 0.0833486863053 |
| Nrf-1 | 0.128398119858 |
| E2a | 0.161346051283 |
| Pur1 | 0.165433943416 |
| Ap-1 | 0.193657210701 |
| Gr | 0.292262463811 |
| Aml1 | 0.357489256791 |
| Myogenin | 0.370547726117 |
| Ap-2rep | 0.442158604448 |
| Sox5 | 0.513017989546 |
| Mafa | 0.612649460032 |
| Pparg | 0.70767586402 |
| TF | P-Value |
| Srf | 3.04311017743e-13 |
| Fxr/rxr-alpha | 0.00189929633743 |
| Stat4 | 0.00253159312556 |
| Sf1 | 0.00281608540835 |
| Hoxc9 | 0.00305820111229 |
| Stat1:stat1 | 0.00463635749024 |
| Rest | 0.0249264791077 |
| Fra1 | 0.028534268049 |
| Nf-y | 0.0400136946424 |
| Hnf4 | 0.0400903766865 |
| Myb | 0.0591210952359 |
| Alpha-cp1 | 0.0708724103187 |
| Esrrb | 0.07449618165 |
| Hnf4, coup | 0.0833486863053 |
| Nrf-1 | 0.128398119858 |
| E2a | 0.161346051283 |
| Pur1 | 0.165433943416 |
| Ap-1 | 0.193657210701 |
| Gr | 0.292262463811 |
| Aml1 | 0.357489256791 |
| Myogenin | 0.370547726117 |
| Ap-2rep | 0.442158604448 |
| Sox5 | 0.513017989546 |
| Mafa | 0.612649460032 |
| Pparg | 0.70767586402 |
| TF | P-Value |
| Srf | 2.88372892741e-48 |
| Creb | 4.55490618875e-34 |
| Hnf4 | 1.52919038906e-14 |
| Creb, atf | 1.00015448614e-12 |
| Atf3 | 9.0638150852e-09 |
| Atf2 | 1.14069130903e-08 |
| Pur1 | 1.19917868768e-07 |
| Sf1 | 1.41329340983e-07 |
| Nrf-1 | 8.09340688379e-07 |
| Smm_as_activator | 8.12672702293e-07 |
| Gabpa | 1.43397094713e-06 |
| Clock:bmal | 1.18652923683e-05 |
| Ap-1 | 3.01807575875e-05 |
| Mzf1 | 9.0908359028e-05 |
| Rest | 0.000192229691471 |
| Atf1 | 0.000197576536952 |
| Elk-1 | 0.000248211697871 |
| Nrse | 0.000281355288212 |
| Usf | 0.000317923059125 |
| Ebox | 0.000504712781209 |
| Ets2 | 0.000546310591494 |
| Ap-2rep | 0.000550709112072 |
| Pparg | 0.000797719419289 |
| Mycn | 0.00106068399233 |
| Mef-2 | 0.00158748066042 |
| TF | P-Value |
| Srf | 7.07576514565e-32 |
| Hnf4 | 7.37324414638e-13 |
| Rest | 7.27008725017e-11 |
| Creb | 6.13757218746e-10 |
| Nrse | 1.27273760189e-09 |
| Nrsf | 9.02513472672e-09 |
| Creb, atf | 1.39733918206e-07 |
| Usf | 1.2652631058e-05 |
| Ets2 | 6.11858063135e-05 |
| Nf-y | 6.63216297022e-05 |
| Atf1 | 6.79191491197e-05 |
| Ap-1 | 0.000118967274906 |
| P63 | 0.000241167202817 |
| Atf2 | 0.000422718233174 |
| Stat1:stat1 | 0.000610223223699 |
| P53 | 0.00064962216608 |
| Ap-2rep | 0.00103896370882 |
| Stat5b (homodimer) | 0.00413833623837 |
| Pur1 | 0.00521446342445 |
| Atf3 | 0.00640122964862 |
| Stat5a (homodimer) | 0.00717150886394 |
| Fxr/rxr-alpha | 0.0104940771863 |
| Mycn | 0.0120006655745 |
| Myc | 0.0131979131376 |
| Clock:bmal | 0.0142201439192 |
| TF | P-Value |
| Hnf4 | 1.6272592686e-78 |
| Srf | 4.33033512878e-61 |
| Creb | 2.04877399603e-42 |
| Creb, atf | 4.98982515143e-21 |
| Gabpa | 9.2285507278e-21 |
| Nrf-1 | 2.1760571658e-20 |
| Pur1 | 1.05000482574e-18 |
| Usf | 2.01519946901e-17 |
| Ets2 | 9.31262996356e-17 |
| Myogenin | 4.77626299902e-16 |
| Ap-2rep | 4.83541875851e-16 |
| Clock:bmal | 3.66354394291e-15 |
| Pparg | 5.14482693546e-15 |
| Sf1 | 1.55669369862e-13 |
| Stat3 | 6.60354379709e-11 |
| Atf2 | 2.18239508632e-10 |
| Gabp | 3.70094442292e-10 |
| Fli1 | 6.57101178254e-10 |
| Elk-1 | 7.44692039287e-10 |
| Mycn | 4.74043171707e-09 |
| Err2 (esrrb) | 6.76637723384e-09 |
| T3r | 1.9887732567e-07 |
| Nf-y | 3.07573875977e-07 |
| Ctcf | 6.33895296985e-07 |
| Nur77 | 1.43855911447e-06 |
| TF | P-Value |
| Gata-1 | 4.28224709421e-05 |
| Bhlhe40_(nb100-1800) | 6.49479388567e-05 |
| Pol2-4h8 | 0.00014043641735 |
| Ctcf_(sc-15914) | 0.00018805920487 |
| Hnf4a | 0.000260971881093 |
| P300_(sc-584) | 0.000327905432402 |
| Ets1 | 0.000383876325718 |
| Znf384_(hpa004051) | 0.000534313309033 |
| Chd2_(ab68301) | 0.000955730859915 |
| Rad21 | 0.00105438960341 |
| Ctcf | 0.00177385744422 |
| Max | 0.00184175685565 |
| Maz_(ab85725) | 0.00199502302416 |
| Smc3_(ab9263) | 0.00302509352656 |
| Gata2_(sc-9008) | 0.00410303574239 |
| Nrsf | 0.00528607054637 |
| C-myc | 0.00749138355306 |
| Gata1_(sc-265) | 0.00816692613606 |
| Zc3h11a_(nb100-74650 | 0.00984601807407 |
| Cebpb | 0.01134234982 |
| C-jun | 0.0137759292745 |
| Tal1_(sc-12984) | 0.0214968778571 |
| P300 | 0.0335074712387 |
| Jund | 0.0363074378776 |
| Corest_(sc-30189) | 0.0377804098704 |
| TF | P-Value |
| Ctcf | 0.00350318188668 |
| Myod_(sc-32758) | 0.0516956142823 |
| Znf384_(hpa004051) | 0.0571747821318 |
| Pol2 | 0.087233526 |
| Myogenin_(sc-12732) | 0.110728008852 |
| Srebf2 | 0.112295348073 |
| Pol2-4h8 | 0.130149150964 |
| P300_(sc-584) | 0.222037380424 |
| Jund | 0.227932723893 |
| Pol2(phosphos2) | 0.228217136073 |
| Mxi1_(af4185) | 0.260287994588 |
| P300 | 0.293279697181 |
| Cry2 | 0.307817573969 |
| Corest_(sc-30189) | 0.314440365387 |
| Hnf4a | 0.318567770784 |
| C-myc | 0.446014607812 |
| Usf2 | 0.449146888611 |
| Tcf12 | 0.456116455101 |
| Smc3_(ab9263) | 0.459011023346 |
| Srebp2 | 0.46108577752 |
| Mafk_(ab50322) | 0.469883097443 |
| Ets1 | 0.494916429623 |
| Usf-1 | 0.512257171411 |
| Znf-mizd-cp1_(ab6576 | 0.533682519063 |
| Zkscan1_(hpa006672) | 0.549293716088 |
| TF | P-Value |
| Pol2(phosphos2) | 5.4675302716e-21 |
| Ctcf_(sc-15914) | 5.52632881615e-20 |
| Gata-1 | 1.07171258859e-19 |
| Pol2 | 2.68022194047e-17 |
| Hnf4a | 1.29851835022e-16 |
| Srebf2 | 3.60297066701e-16 |
| Rad21 | 5.0621002718e-15 |
| Smc3_(ab9263) | 1.59192881558e-13 |
| Pol2-4h8 | 8.06643416174e-12 |
| Ctcf | 1.03488950908e-10 |
| Gata1_(sc-265) | 3.90911933858e-09 |
| Pparg | 5.0847993232e-09 |
| Tcf12 | 7.91770546529e-09 |
| P300_(sc-584) | 1.5495222154e-08 |
| Myod_(sc-32758) | 2.19401343621e-07 |
| Tal1_(sc-12984) | 2.20155738668e-07 |
| P300 | 2.27367711318e-07 |
| Srebf1 | 2.65561534645e-07 |
| Gata2_(sc-9008) | 4.61944240156e-07 |
| Corest_(sc-30189) | 5.08591773419e-07 |
| Usf2 | 1.50439075921e-06 |
| Mafk_(ab50322) | 3.79888828625e-06 |
| Znf384_(hpa004051) | 3.94254953372e-06 |
| Ubf_(sc-13125) | 4.75552931336e-06 |
| Cebpb | 4.83007652753e-05 |
| TF | P-Value |
| Pol2(phosphos2) | 5.24753084726e-13 |
| Ctcf_(sc-15914) | 2.00484805581e-09 |
| Smc3_(ab9263) | 8.95178591849e-09 |
| Rad21 | 6.55126453263e-08 |
| Srebf2 | 1.40632833065e-07 |
| Pol2 | 1.97985235735e-06 |
| Ctcf | 2.06730042934e-06 |
| Gata-1 | 2.56853095784e-06 |
| Hnf4a | 7.65282435323e-06 |
| Srebf1 | 4.61055712473e-05 |
| Zc3h11a_(nb100-74650 | 0.000114188485713 |
| Corest_(sc-30189) | 0.000473036077653 |
| Jund | 0.000496020528574 |
| P300_(sc-584) | 0.00064235319789 |
| Gata1_(sc-265) | 0.000981124486987 |
| Ets1 | 0.00100664459455 |
| Pol2-4h8 | 0.00108306363442 |
| Ubf_(sc-13125) | 0.00123496338462 |
| Tcf12 | 0.00164233211386 |
| Myod_(sc-32758) | 0.00183527707491 |
| Tal1_(sc-12984) | 0.00362988329344 |
| Bhlhe40_(nb100-1800) | 0.00447365711016 |
| Nelfe | 0.00700100273332 |
| Znf-mizd-cp1_(ab6576 | 0.0100105562049 |
| Usf2 | 0.0135020471664 |
| TF | P-Value |
| Pol2(phosphos2) | 0.000158296845945 |
| Pol2 | 0.000699522905197 |
| Gata-1 | 0.00606692900425 |
| Gata1_(sc-265) | 0.0093547457303 |
| Rad21 | 0.0130970149227 |
| Smc3_(ab9263) | 0.0152038375819 |
| Ctcf_(sc-15914) | 0.0211426270942 |
| Pol2-4h8 | 0.0228923846071 |
| Mafk_(ab50322) | 0.0282741953868 |
| Hnf4a | 0.0288395095482 |
| Znf384_(hpa004051) | 0.0373850682252 |
| Ctcf | 0.0410482028293 |
| Tcf12 | 0.0575933260708 |
| Gata2_(sc-9008) | 0.0590249718451 |
| Myogenin_(sc-12732) | 0.0716899694773 |
| Tal1_(sc-12984) | 0.0806067385085 |
| Fli1_(sc-356) | 0.0827113274323 |
| Myod_(sc-32758) | 0.0909430565142 |
| Chd1_(nb100-60411) | 0.103998770878 |
| C-jun | 0.124001503198 |
| E2f4 | 0.125618456298 |
| Usf-1 | 0.146305696325 |
| Hcfc1_(nb100-68209) | 0.154137810898 |
| Fosl1_(sc-605) | 0.167150105649 |
| Pparg | 0.170125340691 |
| TF | P-Value |
| Pparg | 2.14707806259e-10 |
| Ctcf_(sc-15914) | 9.20941871574e-08 |
| P300_(sc-584) | 1.32257187523e-07 |
| Pol2 | 2.32798735962e-07 |
| Ets1 | 4.75645352272e-07 |
| Rad21 | 1.06024801011e-06 |
| Bhlhe40_(nb100-1800) | 3.89294042892e-06 |
| Tbp | 5.84609421874e-06 |
| Mafk_(ab50322) | 6.89212856877e-06 |
| Gata-1 | 8.14553140168e-06 |
| Srebf2 | 8.71907530687e-06 |
| Max | 1.46931236074e-05 |
| Hnf4a | 1.56749867723e-05 |
| P300 | 1.91029420358e-05 |
| C-myc | 4.4217605152e-05 |
| Pol2-4h8 | 7.49903720935e-05 |
| Smc3_(ab9263) | 7.95448398001e-05 |
| Ctcf | 0.000107566913308 |
| Cry1 | 0.000220372709967 |
| Tcf12 | 0.000222300652763 |
| Znf384_(hpa004051) | 0.000377395820499 |
| Sin3a_(nb600-1263) | 0.000644131907193 |
| Corest_(sc-30189) | 0.000686932061422 |
| Nelfe | 0.00110332544868 |
| Myod_(sc-32758) | 0.00123826834409 |
| TF | P-Value |
| Gata-1 | 0.0001490406539 |
| P300 | 0.000439722112881 |
| Pparg | 0.000567729978161 |
| P300_(sc-584) | 0.00125060073207 |
| Ctcf | 0.00197018368821 |
| Ets1 | 0.00566813061724 |
| Pol2 | 0.00703895807927 |
| Corest_(sc-30189) | 0.00728322371971 |
| Hnf4a | 0.00856435571011 |
| Rad21 | 0.00876193636757 |
| Ctcf_(sc-15914) | 0.00876193636757 |
| Myod_(sc-32758) | 0.00986941088541 |
| Mafk_(ab50322) | 0.0135560732743 |
| Tbp | 0.0142475218254 |
| Pol2-4h8 | 0.0145020028924 |
| Smc3_(ab9263) | 0.019802839581 |
| Sin3a_(nb600-1263) | 0.0269838178539 |
| Tcf12 | 0.0290280697087 |
| Nelfe | 0.0303721339003 |
| Srebf2 | 0.0337281981023 |
| Usf-1 | 0.0338058941229 |
| Tal1_(sc-12984) | 0.035494922926 |
| Znf384_(hpa004051) | 0.0362648962641 |
| Bhlhe40_(nb100-1800) | 0.0500671088076 |
| Max | 0.0543273972398 |
| TF | P-Value |
| Pol2 | 0.0181556078151 |
| Ubf_(sc-13125) | 0.0209488937879 |
| Max | 0.0305198549292 |
| Nelfe | 0.0351996720596 |
| Tbp | 0.0539271141913 |
| Tcf3_(sc-349) | 0.0650379069192 |
| Srebf2 | 0.075089834631 |
| Hcfc1_(nb100-68209) | 0.0953218813131 |
| P300_(sc-584) | 0.0963330665473 |
| Cry2 | 0.102927379903 |
| Bhlhe40_(nb100-1800) | 0.103294576103 |
| Ctcf | 0.103970456831 |
| Clock | 0.104550244239 |
| Gata-1 | 0.118555907943 |
| Sin3a_(nb600-1263) | 0.125106768644 |
| Tal1_(sc-12984) | 0.133294930876 |
| Ctcf_(sc-15914) | 0.152368848785 |
| Nrsf | 0.157711025457 |
| Smc3_(ab9263) | 0.157911100072 |
| Pol2(phosphos2) | 0.180644612844 |
| Gcn5 | 0.191639524664 |
| Corest_(sc-30189) | 0.201472446136 |
| Gata1_(sc-265) | 0.211378450901 |
| Znf384_(hpa004051) | 0.214557277977 |
| Mxi1_(af4185) | 0.219553315761 |
| TF | P-Value |
| Srebf2 | 4.48619454617e-10 |
| Pol2 | 8.13195452092e-10 |
| Sin3a_(nb600-1263) | 6.34600663038e-07 |
| Mafk_(ab50322) | 3.11962798404e-06 |
| Mxi1_(af4185) | 4.02446289716e-06 |
| Pol2(phosphos2) | 9.39068968493e-06 |
| Gcn5 | 1.81928345951e-05 |
| Myod_(sc-32758) | 2.53071371354e-05 |
| Fli1_(sc-356) | 5.66201513353e-05 |
| Srebf1 | 0.000133223774985 |
| Max | 0.00018770503513 |
| C-myc | 0.000220574957425 |
| Tcf12 | 0.000229653608855 |
| Pol2-4h8 | 0.000314864410262 |
| P300 | 0.000378389928476 |
| Tbp | 0.000389928999666 |
| Hcfc1_(nb100-68209) | 0.000437524606476 |
| Ctcf | 0.000557621285522 |
| Ctcf_(sc-15914) | 0.000612447395061 |
| Nrf2 | 0.00174805663463 |
| Rad21 | 0.00249599026336 |
| Smc3_(ab9263) | 0.0036908697511 |
| Gata2_(sc-9008) | 0.00739425469161 |
| Bhlhe40_(nb100-1800) | 0.00831630195226 |
| E2f4 | 0.0103726867673 |
| TF | P-Value |
| Srebf2 | 4.29529462775e-12 |
| Pol2 | 3.36579927743e-10 |
| Mafk_(ab50322) | 7.1663744379e-08 |
| Myod_(sc-32758) | 1.87378082622e-06 |
| Pol2(phosphos2) | 4.81342834918e-05 |
| Srebf1 | 7.6611729426e-05 |
| Fli1_(sc-356) | 7.887536051e-05 |
| Tcf12 | 0.000168983198038 |
| Sin3a_(nb600-1263) | 0.000212525858035 |
| Ctcf | 0.000229580002109 |
| Rad21 | 0.000395985073067 |
| Max | 0.000440636312879 |
| Bhlhe40_(nb100-1800) | 0.000448731420884 |
| Ctcf_(sc-15914) | 0.000523319847367 |
| Gcn5 | 0.00125034018854 |
| Smc3_(ab9263) | 0.0013857233406 |
| Mxi1_(af4185) | 0.00158305577208 |
| E2f4 | 0.00198195622985 |
| Usf2 | 0.00206159983903 |
| P300 | 0.00241311753657 |
| Tcf3_(sc-349) | 0.00307005562366 |
| Gata1_(sc-265) | 0.00512795423605 |
| Tbp | 0.00599788079217 |
| Ets1 | 0.00646986076342 |
| C-myc | 0.00868161339468 |
| TF | P-Value |
| Pol2 | 3.67518932469e-10 |
| Srebf2 | 1.16730145288e-09 |
| Mafk_(ab50322) | 2.20559587534e-08 |
| Myod_(sc-32758) | 1.14398084339e-06 |
| Fli1_(sc-356) | 1.56903486744e-05 |
| E2f4 | 2.89042184783e-05 |
| Ets1 | 3.7332683899e-05 |
| Tcf12 | 4.87790477363e-05 |
| Bhlhe40_(nb100-1800) | 9.09540130422e-05 |
| Srebf1 | 0.000164480341002 |
| Rad21 | 0.000310654239381 |
| Usf2 | 0.000429853593346 |
| Max | 0.000522617470218 |
| Ctcf | 0.000558359180515 |
| P300_(sc-584) | 0.000855913946376 |
| Ctcf_(sc-15914) | 0.000943040190014 |
| Pol2(phosphos2) | 0.00165614962004 |
| Mxi1_(af4185) | 0.00186630606585 |
| Smc3_(ab9263) | 0.00252665899896 |
| Sin3a_(nb600-1263) | 0.0044922125423 |
| Gcn5 | 0.00608896578312 |
| Pparg | 0.00760489411573 |
| Tal1_(sc-12984) | 0.00954035733693 |
| Cebpb | 0.0114029265903 |
| Gata2_(sc-9008) | 0.0135565529222 |
| TF | P-Value |
| Pol2 | 2.66118336405e-10 |
| Sin3a_(nb600-1263) | 3.22062385985e-05 |
| Pol2-4h8 | 0.000206823762453 |
| Mxi1_(af4185) | 0.00227267984681 |
| Hcfc1_(nb100-68209) | 0.00419496872498 |
| Nelfe | 0.00786066008918 |
| Ctcf | 0.00900226745858 |
| Cry1 | 0.00945178623978 |
| Maz_(ab85725) | 0.011258266065 |
| Myod_(sc-32758) | 0.0226818161385 |
| Chd2_(ab68301) | 0.025119835586 |
| Bmal | 0.0310049860807 |
| Gata1_(sc-265) | 0.0395998333857 |
| Gcn5 | 0.0423414503346 |
| Tcf12 | 0.0600964708838 |
| Hnf4a | 0.0613521739497 |
| Bhlhe40_(nb100-1800) | 0.066814516322 |
| E2f4 | 0.0674910761958 |
| Znf-mizd-cp1_(ab6576 | 0.0737056962124 |
| Tbp | 0.0798170160737 |
| Cebpb | 0.0815145495178 |
| Mafk_(ab50322) | 0.0815895729486 |
| Tcf3_(sc-349) | 0.0985717033866 |
| Ets1 | 0.10760376102 |
| Tal1_(sc-12984) | 0.113003478292 |
| TF | P-Value |
| Pol2 | 8.64355613528e-08 |
| Gata2_(sc-9008) | 8.06315703862e-05 |
| Pol2-4h8 | 0.000162044647135 |
| Myod_(sc-32758) | 0.00236166547343 |
| Sin3a_(nb600-1263) | 0.00348071444014 |
| Pol2(phosphos2) | 0.00354685138496 |
| Pparg | 0.00365564237785 |
| Tcf12 | 0.00404617058259 |
| Hcfc1_(nb100-68209) | 0.00419496872498 |
| Gata1_(sc-265) | 0.00698028599286 |
| Usf2 | 0.00842522401203 |
| Gcn5 | 0.0135946768781 |
| Fosl1_(sc-605) | 0.0245613438473 |
| Cbp | 0.0270303373024 |
| Max | 0.0324855570517 |
| Mafk_(ab50322) | 0.0491898808737 |
| Tal1_(sc-12984) | 0.0495238827731 |
| Ubf_(sc-13125) | 0.0499285222923 |
| Usf-1 | 0.0516667122026 |
| Jund | 0.0572866418934 |
| Znf-mizd-cp1_(ab6576 | 0.0674593179936 |
| Cebpb | 0.0917817714119 |
| P500 | 0.0926711753827 |
| Ctcf | 0.111076935487 |
| Tbp | 0.122009369079 |
| TF | P-Value |
| None |
| TF | P-Value |
| None |
| TF | P-Value |
| Ctcf_(sc-15914) | 2.83471838471e-22 |
| Mafk_(ab50322) | 4.8898808406e-21 |
| Rad21 | 2.74375157228e-20 |
| Bhlhe40_(nb100-1800) | 2.66196834976e-19 |
| Pol2-4h8 | 6.98617204149e-17 |
| Ets1 | 5.82887588474e-16 |
| Gata-1 | 1.45286187142e-15 |
| Usf2 | 1.59448383414e-15 |
| Smc3_(ab9263) | 3.83075155004e-15 |
| Srebf2 | 1.12293223015e-13 |
| Hnf4a | 3.12201255712e-13 |
| P300_(sc-584) | 2.29840989105e-12 |
| Ctcf | 5.97798799535e-12 |
| Maz_(ab85725) | 7.14358063355e-12 |
| Corest_(sc-30189) | 1.08176175201e-11 |
| Pparg | 2.07904957197e-11 |
| Myod_(sc-32758) | 2.91338906125e-11 |
| Pol2 | 1.36260133153e-10 |
| Pol2(phosphos2) | 2.69446412078e-10 |
| Znf384_(hpa004051) | 3.81400763859e-09 |
| Srebf1 | 1.36123871329e-08 |
| Chd2_(ab68301) | 2.67687599122e-08 |
| Tal1_(sc-12984) | 7.31593792624e-08 |
| Tcf12 | 8.43271045581e-08 |
| Cebpb | 1.21029754997e-07 |
| TF | P-Value |
| Ctcf_(sc-15914) | 6.67036045894e-26 |
| Rad21 | 3.63726106938e-23 |
| Mafk_(ab50322) | 2.74396310346e-19 |
| Bhlhe40_(nb100-1800) | 8.66080279188e-19 |
| Smc3_(ab9263) | 7.49857555133e-18 |
| Ctcf | 7.95562844456e-15 |
| Gata-1 | 9.23362606156e-15 |
| Usf2 | 9.61701873671e-15 |
| Ets1 | 1.87571051043e-14 |
| Pol2-4h8 | 1.88423205611e-14 |
| Pol2 | 2.10129300422e-14 |
| Srebf2 | 9.89091918169e-14 |
| Myod_(sc-32758) | 1.23282666173e-13 |
| Corest_(sc-30189) | 4.32541017505e-12 |
| Hnf4a | 8.0393228572e-12 |
| Maz_(ab85725) | 2.48404220902e-11 |
| Pparg | 4.90262177357e-10 |
| P300_(sc-584) | 1.42164761623e-09 |
| Pol2(phosphos2) | 1.20210527394e-08 |
| Tcf12 | 1.45725716123e-08 |
| Max | 4.06097862783e-08 |
| Myogenin_(sc-12732) | 1.2402264895e-07 |
| Cebpb | 1.69368329171e-07 |
| Chd2_(ab68301) | 1.77030414278e-07 |
| Znf384_(hpa004051) | 3.08059855867e-07 |
| TF | P-Value |
| Ctcf_(sc-15914) | 1.19098050267e-22 |
| Rad21 | 1.87995354056e-21 |
| Mafk_(ab50322) | 3.29731368052e-17 |
| Bhlhe40_(nb100-1800) | 2.446797135e-16 |
| Smc3_(ab9263) | 1.78122125641e-15 |
| Gata-1 | 5.84208712134e-14 |
| Pol2 | 2.20980082468e-13 |
| Myod_(sc-32758) | 4.04129613605e-13 |
| Usf2 | 2.48131298074e-12 |
| Srebf2 | 3.95160144637e-12 |
| Ctcf | 9.06505660994e-12 |
| Ets1 | 4.86484167041e-11 |
| Tcf12 | 1.81186542736e-10 |
| Hnf4a | 3.14442812446e-10 |
| Pol2-4h8 | 9.98979865903e-10 |
| Maz_(ab85725) | 1.50854632708e-09 |
| Corest_(sc-30189) | 1.86334188709e-09 |
| P300_(sc-584) | 4.93709976548e-08 |
| Pol2(phosphos2) | 7.95538696323e-08 |
| Cebpb | 2.75144446945e-07 |
| Myogenin_(sc-12732) | 5.20184624618e-07 |
| Srebf1 | 6.67781620225e-07 |
| Chd2_(ab68301) | 1.12705394662e-06 |
| Max | 1.16890766708e-06 |
| Pparg | 2.4590593406e-06 |
| TF | P-Value |
| Pol2 | 1.23425475203e-13 |
| Sin3a_(nb600-1263) | 1.97900468406e-05 |
| P300 | 7.69765726777e-05 |
| Hnf4a | 0.000275768428343 |
| Pparg | 0.000368326111289 |
| Mxi1_(af4185) | 0.00132970972061 |
| Max | 0.00147637987837 |
| Gcn5 | 0.00176287778077 |
| Cry1 | 0.00295346086134 |
| Srebf1 | 0.0055251413478 |
| Pol2(phosphos2) | 0.00553055251434 |
| Fosl1_(sc-605) | 0.00570669572523 |
| C-myc | 0.00674765915423 |
| Ctcf | 0.00708642313976 |
| Srebp1 | 0.0170303620562 |
| Gata-1 | 0.017199126055 |
| Srebf2 | 0.0172723666726 |
| Mafk_(ab50322) | 0.0173373257822 |
| Myogenin_(sc-12732) | 0.019769889303 |
| Zc3h11a_(nb100-74650 | 0.0231347682813 |
| Hcfc1_(nb100-68209) | 0.0264790150164 |
| P300_(sc-584) | 0.030703490786 |
| Usf2 | 0.0335763259894 |
| Tbp | 0.0361929782069 |
| Gata2_(sc-9008) | 0.0470750732523 |
| TF | P-Value |
| Pol2 | 2.51990482037e-05 |
| Srebf1 | 0.000250723662083 |
| Ctcf | 0.000336094816834 |
| Sin3a_(nb600-1263) | 0.0040169049886 |
| Gcn5 | 0.00454353907933 |
| Hcfc1_(nb100-68209) | 0.00691094810839 |
| Gata2_(sc-9008) | 0.00910120773579 |
| Tcf3_(sc-349) | 0.0135347065586 |
| Gata1_(sc-265) | 0.0146330171541 |
| Pol2-4h8 | 0.0238152524379 |
| Pparg | 0.0259374125538 |
| Ets1 | 0.0263436041762 |
| Maz_(ab85725) | 0.0387670219586 |
| Srebf2 | 0.0427621976183 |
| Nrsf | 0.058023885001 |
| Jund | 0.0658972241502 |
| Nrf2 | 0.0726087221375 |
| Mxi1_(af4185) | 0.074186460834 |
| C-jun | 0.0792039864145 |
| Max | 0.089949442077 |
| Tcf12 | 0.0908488134729 |
| Myod_(sc-32758) | 0.1010171141 |
| P300_(sc-584) | 0.106463019552 |
| Myogenin_(sc-12732) | 0.117943101217 |
| Nelfe | 0.13489054448 |
| TF | P-Value |
| Pol2 | 1.28249433707e-08 |
| Gata-1 | 1.17343234106e-05 |
| Mxi1_(af4185) | 0.000998262708446 |
| Hcfc1_(nb100-68209) | 0.00109336396499 |
| Pol2-4h8 | 0.00121470028688 |
| Chd2_(ab68301) | 0.00132909530105 |
| Sin3a_(nb600-1263) | 0.00259186705841 |
| Ctcf | 0.00308450229077 |
| Nelfe | 0.00341810198926 |
| E2f4 | 0.00357846429825 |
| C-myc | 0.00372658529517 |
| Maz_(ab85725) | 0.00702021611013 |
| P300_(sc-584) | 0.00727029690507 |
| Tcf12 | 0.00784429354796 |
| Hnf4a | 0.0156622926131 |
| Gcn5 | 0.018447200654 |
| Mafk_(ab50322) | 0.0199696730184 |
| Corest_(sc-30189) | 0.0261427682425 |
| C-myb_(sc-7874) | 0.0289267831891 |
| Tcf3_(sc-349) | 0.0309117419948 |
| P300 | 0.0309566631448 |
| Pparg | 0.0327458987755 |
| Bhlhe40_(nb100-1800) | 0.0375487752286 |
| Max | 0.0488141054286 |
| Tal1_(sc-12984) | 0.0530572212756 |
| TF | P-Value |
| Pol2 | 1.17464605509e-07 |
| Ctcf | 2.51094272171e-06 |
| Sin3a_(nb600-1263) | 3.23734035915e-05 |
| Pparg | 0.000119658901732 |
| Gcn5 | 0.000247174707387 |
| Pol2-4h8 | 0.000675720156537 |
| Hnf4a | 0.000840989344074 |
| Srebf1 | 0.00430272343983 |
| Hcfc1_(nb100-68209) | 0.00565015830684 |
| Maz_(ab85725) | 0.0113759726958 |
| Myod_(sc-32758) | 0.0122598999326 |
| Srebf2 | 0.0123522668817 |
| Mxi1_(af4185) | 0.0139426830268 |
| Srebp1 | 0.0202542652299 |
| Zc3h11a_(nb100-74650 | 0.0238933915706 |
| Tcf3_(sc-349) | 0.0271951534884 |
| Mafk_(ab50322) | 0.0313718509127 |
| Max | 0.0332806840061 |
| Nelfe | 0.0371690706583 |
| Gata1_(sc-265) | 0.0418224806127 |
| Nrsf | 0.0475328797665 |
| Cry1 | 0.0531080663905 |
| Znf384_(hpa004051) | 0.0670244045049 |
| Tbp | 0.0711569932671 |
| Ets1 | 0.0796244557569 |
| TF | P-Value |
| Pol2 | 1.30531511315e-05 |
| Pol2-4h8 | 6.37533228203e-05 |
| E2f4 | 0.00904205978001 |
| Sin3a_(nb600-1263) | 0.0108579670946 |
| Hcfc1_(nb100-68209) | 0.0156247559469 |
| Gata1_(sc-265) | 0.0451041692761 |
| Ctcf | 0.0571293733292 |
| Tal1_(sc-12984) | 0.0613730348761 |
| Myogenin_(sc-12732) | 0.067278496741 |
| Max | 0.0708977322246 |
| Nrf2 | 0.0841765643048 |
| Srebp1 | 0.0899712817283 |
| Znf384_(hpa004051) | 0.0908907218056 |
| Ets1 | 0.105618686269 |
| Gcn5 | 0.106421730384 |
| Tcf12 | 0.114284071649 |
| Smc3_(ab9263) | 0.159630746048 |
| Gata2_(sc-9008) | 0.176953166522 |
| Usf-1 | 0.178510778178 |
| P300_(sc-584) | 0.182687593684 |
| Gata-1 | 0.184398773628 |
| Srebf2 | 0.204314309625 |
| Rad21 | 0.216912980413 |
| P300 | 0.226787055725 |
| Znf-mizd-cp1_(ab6576 | 0.252969635606 |
| TF | P-Value |
| Gata-1 | 2.40654467695e-20 |
| Ctcf_(sc-15914) | 3.31300190158e-14 |
| Pol2 | 2.20643366772e-13 |
| Max | 1.48018471705e-12 |
| Bhlhe40_(nb100-1800) | 3.21420481312e-11 |
| Znf384_(hpa004051) | 7.53980481534e-11 |
| P300 | 9.10440078815e-11 |
| Usf2 | 7.91524845235e-10 |
| Ctcf | 1.81766764257e-09 |
| Maz_(ab85725) | 5.00159471235e-09 |
| Rad21 | 6.88894768993e-09 |
| C-myc | 7.09484402028e-09 |
| Sin3a_(nb600-1263) | 5.39687323347e-08 |
| Chd2_(ab68301) | 8.90520420959e-08 |
| Pol2-4h8 | 1.97111906851e-07 |
| Corest_(sc-30189) | 2.42457105087e-07 |
| Srebf2 | 5.90427992586e-07 |
| Hnf4a | 6.40242512566e-07 |
| Pparg | 1.04216986459e-06 |
| Gcn5 | 1.10733331703e-06 |
| Srebp1 | 1.92725524512e-06 |
| Tbp | 2.26400227914e-06 |
| Mxi1_(af4185) | 3.41082601471e-06 |
| Nelfe | 4.73637169932e-06 |
| Zkscan1_(hpa006672) | 7.04848287817e-06 |
| TF | P-Value |
| Yy1 | 1.11018717961e-07 |
| Nrf-1 | 6.14037167032e-07 |
| Sox5 | 8.06501749129e-07 |
| Math1 | 0.000248580203582 |
| Nf-y | 0.000996820343164 |
| Heb | 0.0012456120215 |
| Nrf-2 | 0.00229297854545 |
| Hbp1 | 0.00239132736557 |
| Lbp9 (tcfcp211) | 0.00241038921064 |
| Srebp-1 | 0.00378320107433 |
| Nkx6-2 | 0.00526100866463 |
| Nrsf | 0.00554503621754 |
| Maf | 0.00597144997214 |
| Nrse | 0.00597227942528 |
| Nobox | 0.010617195517 |
| Tbx15 | 0.014560681397 |
| Hnf3a | 0.0204029872551 |
| Usf | 0.0230873384012 |
| Nkx2-5 | 0.0316093086236 |
| Dmrt7 | 0.0339639345435 |
| Rest | 0.0418775128567 |
| Hfh8 (foxf1a) | 0.0505108404994 |
| Egr-3 | 0.0505108404994 |
| Arnt | 0.0512609352204 |
| Gsh2 | 0.0543293625477 |
| TF | P-Value |
| Max | 3.10129492822e-05 |
| Pol2 | 5.53925157397e-05 |
| Ctcf_(sc-15914) | 7.36917070673e-05 |
| Pol2-4h8 | 0.000321979532712 |
| Gcn5 | 0.000540620384426 |
| Srebf2 | 0.00128874534663 |
| Ctcf | 0.00147923796472 |
| C-myc | 0.00185617100655 |
| Zkscan1_(hpa006672) | 0.00259958516282 |
| Chd2_(ab68301) | 0.00316906640637 |
| Sin3a_(nb600-1263) | 0.00411685287345 |
| Rad21 | 0.00455301578556 |
| Corest_(sc-30189) | 0.00502361740989 |
| Bhlhe40_(nb100-1800) | 0.00528059212971 |
| Gata-1 | 0.00606478403976 |
| Mxi1_(af4185) | 0.00647312598038 |
| Gata2_(sc-9008) | 0.00916306929889 |
| Maz_(ab85725) | 0.0103019217128 |
| Usf2 | 0.0104746694492 |
| Tbp | 0.0122531075053 |
| Hcfc1_(nb100-68209) | 0.0142930750809 |
| Bmal1 | 0.0162996398468 |
| C-myb_(sc-7874) | 0.0242311817057 |
| P300 | 0.0286851684441 |
| Tal1_(sc-12984) | 0.0309600693814 |
| TF | P-Value |
| Pol2 | 0.000346095896747 |
| Mxi1_(af4185) | 0.00125290708997 |
| Hnf4a | 0.00718143425578 |
| Sin3a_(nb600-1263) | 0.011157327853 |
| Pparg | 0.0179160823548 |
| Gata1_(sc-265) | 0.0239199385411 |
| Zkscan1_(hpa006672) | 0.029541817089 |
| C-myb_(sc-7874) | 0.0368653746086 |
| Bhlhe40_(nb100-1800) | 0.0375037351658 |
| Fli1_(sc-356) | 0.072460239744 |
| Ctcf_(sc-15914) | 0.0785670795186 |
| Gata-1 | 0.0834869153161 |
| Hcfc1_(nb100-68209) | 0.0849964369239 |
| Usf2 | 0.0881520170277 |
| C-myc | 0.0953947686207 |
| Per1 | 0.106996233908 |
| Max | 0.112788424576 |
| Ctcf | 0.113325276698 |
| Nrf2 | 0.11523105483 |
| Chd2_(ab68301) | 0.124049746273 |
| Chd1_(nb100-60411) | 0.124954739955 |
| Bmal | 0.1251924647 |
| Cry1 | 0.127000531725 |
| Pol2(phosphos2) | 0.13617868302 |
| Znf384_(hpa004051) | 0.162055989678 |
| TF | P-Value |
| Gata-1 | 2.53317131559e-66 |
| Ctcf_(sc-15914) | 8.06924941155e-60 |
| Bhlhe40_(nb100-1800) | 2.95919263941e-53 |
| Max | 2.33742398391e-49 |
| Rad21 | 4.94427107429e-41 |
| P300 | 3.68782767128e-40 |
| Maz_(ab85725) | 1.23322032749e-36 |
| Pol2-4h8 | 1.66961247347e-34 |
| C-myc | 2.24963149128e-32 |
| Ctcf | 4.56655573195e-31 |
| Sin3a_(nb600-1263) | 1.47040917797e-30 |
| Mxi1_(af4185) | 2.73251557035e-30 |
| Corest_(sc-30189) | 2.01917952379e-29 |
| Ets1 | 5.77399402878e-29 |
| Pparg | 6.4768326218e-28 |
| Znf384_(hpa004051) | 8.07270962101e-28 |
| Usf2 | 2.77371416315e-27 |
| Hnf4a | 8.07618936619e-27 |
| Tbp | 3.25610942047e-26 |
| Nrsf | 4.63317988121e-26 |
| Smc3_(ab9263) | 8.94590924226e-25 |
| Mafk_(ab50322) | 2.83867799724e-23 |
| Srebf2 | 5.40873469398e-22 |
| Chd2_(ab68301) | 1.38992305637e-21 |
| Pol2 | 8.25776358764e-20 |
| TF | P-Value |
| Gata-1 | 1.15734689469e-53 |
| Ctcf_(sc-15914) | 2.18098727206e-44 |
| Rad21 | 1.92328289477e-30 |
| Max | 7.85609484241e-29 |
| Bhlhe40_(nb100-1800) | 1.20468747168e-28 |
| Maz_(ab85725) | 2.51226851236e-28 |
| Ctcf | 2.87952889401e-27 |
| Znf384_(hpa004051) | 1.34917682959e-26 |
| Pol2-4h8 | 6.01346829979e-25 |
| P300 | 1.21486896299e-23 |
| Corest_(sc-30189) | 5.22498397166e-21 |
| Sin3a_(nb600-1263) | 8.43797238182e-20 |
| Mxi1_(af4185) | 1.48051060068e-19 |
| C-myc | 3.6744953779e-19 |
| Hnf4a | 7.44121073009e-19 |
| Usf2 | 1.26885560589e-18 |
| Tbp | 3.29985386689e-18 |
| Smc3_(ab9263) | 4.00964277101e-18 |
| Ets1 | 2.2985882688e-17 |
| Mafk_(ab50322) | 3.11744007615e-17 |
| Nrsf | 2.26694306465e-16 |
| Gata1_(sc-265) | 8.31635315082e-16 |
| Pol2 | 7.37284353279e-15 |
| Gcn5 | 6.89420996446e-14 |
| Usf-1 | 8.06267912778e-14 |
| TF | P-Value |
| Lbp9 (tcfcp211) | 1.86697733228e-05 |
| Hbp1 | 0.000367897889728 |
| Nf-y | 0.00289552993506 |
| Maf | 0.00372178541869 |
| Nrf-2 | 0.0046611648197 |
| Hfh8 (foxf1a) | 0.0152632306263 |
| Nkx2-5 | 0.0181178582387 |
| Hnf3a | 0.0246455368876 |
| Nrsf | 0.0261010584017 |
| Yy1 | 0.0279547728573 |
| Spz1 | 0.0352524288498 |
| Err2 (esrrb) | 0.0353114625528 |
| Sox5 | 0.039219990854 |
| C-myb | 0.0392231746113 |
| Hnf4, coup | 0.0436325694441 |
| Nrse | 0.0437794602695 |
| Srebp-1 | 0.0468751995463 |
| Tcfcp2l1 | 0.0668885527479 |
| Heb | 0.0760987778906 |
| Zic1 | 0.0859982790319 |
| Lef1, tcf1 | 0.106090470937 |
| Arnt | 0.122214170156 |
| Rest | 0.127185440749 |
| Sf1 | 0.129519233317 |
| Stat5b (homodimer) | 0.132941569145 |
| TF | P-Value |
| Ctcf_(sc-15914) | 4.11457913093e-13 |
| Znf384_(hpa004051) | 8.49454806215e-10 |
| Gata-1 | 2.74144068977e-09 |
| Rad21 | 5.63130506393e-09 |
| Ctcf | 7.86778094392e-08 |
| Bhlhe40_(nb100-1800) | 8.79142998243e-08 |
| Smc3_(ab9263) | 2.28165259877e-06 |
| Max | 2.29086792321e-06 |
| Gata1_(sc-265) | 3.45452873913e-06 |
| Mxi1_(af4185) | 3.89384611562e-06 |
| Maz_(ab85725) | 4.69523861582e-06 |
| Pol2-4h8 | 8.60695822021e-06 |
| Sin3a_(nb600-1263) | 9.88851344105e-06 |
| Chd2_(ab68301) | 1.97588926038e-05 |
| C-myc | 2.38788862011e-05 |
| Corest_(sc-30189) | 4.35771988657e-05 |
| Pol2 | 8.08743061573e-05 |
| P300 | 0.000120690265287 |
| Myod_(sc-32758) | 0.000123015308681 |
| Hnf4a | 0.000193766752478 |
| Pol2(phosphos2) | 0.000195691568859 |
| Gata2_(sc-9008) | 0.00027731800091 |
| Usf2 | 0.000324882434222 |
| Tal1_(sc-12984) | 0.000370626871634 |
| Tbp | 0.000420623747632 |
| TF | P-Value |
| Hnf4 | 4.96419924455e-31 |
| Ap-2rep | 6.20005786247e-13 |
| Ets2 | 2.642162709e-11 |
| Pparg | 8.62610111311e-08 |
| Pur1 | 1.59107393146e-06 |
| Stat6 | 9.68225076844e-06 |
| Myogenin | 1.41590827959e-05 |
| Lbp9 (tcfcp211) | 1.72955862727e-05 |
| Creb | 0.000148161657675 |
| Nf-1 | 0.000750946854933 |
| Math1 | 0.00120521521796 |
| Hnf4, coup | 0.00135956437045 |
| Ctcf | 0.0018089932192 |
| Mycn | 0.00207493823153 |
| Atf3 | 0.0021136021312 |
| Srf | 0.00527816325699 |
| Atf1 | 0.0059901392748 |
| Hen1 | 0.00715587629101 |
| Pax-6 | 0.00873475552345 |
| Smad3 | 0.00969765601678 |
| C/ebpdelta | 0.0100628587248 |
| Myb | 0.0105702514698 |
| Atf2 | 0.0116809135851 |
| Stat3 | 0.0130369890383 |
| P63 | 0.0135564653447 |
| TF | P-Value |
| Hnf4 | 2.28182896371e-11 |
| Ets2 | 4.49895504959e-07 |
| Creb | 4.40778487499e-06 |
| Ap-2rep | 5.1363234451e-06 |
| Pparg | 5.62681626996e-05 |
| Pur1 | 6.48227826536e-05 |
| Nf-1 | 0.000583997684887 |
| Stat6 | 0.000908407687271 |
| Atf2:c-jun | 0.00121456801775 |
| Atf2 | 0.00262537699196 |
| C-ets-1 | 0.00697383027272 |
| Hoxd3 | 0.0116477611094 |
| Tata | 0.0129335529926 |
| Atf1 | 0.017623688011 |
| Mox1 | 0.0180600147871 |
| Rfx1 | 0.0290480802556 |
| T3r | 0.032544083748 |
| Hoxa2 | 0.0357938646533 |
| Tal1::gata1 | 0.0357938646533 |
| Creb, atf | 0.0369318594916 |
| Myb | 0.0408297302174 |
| Deltaef1 | 0.0469488535558 |
| Hoxa1 | 0.0482644746911 |
| Nf-kappab | 0.0532019034237 |
| Nfat3 | 0.0618252122484 |
| TF | P-Value |
| Pol2-4h8 | 6.02956278354e-16 |
| Ctcf_(sc-15914) | 1.85657282174e-10 |
| Hnf4a | 7.96569051959e-09 |
| Gata-1 | 8.40429488631e-09 |
| Rad21 | 1.91215457912e-08 |
| Ets1 | 4.61860688553e-08 |
| Bhlhe40_(nb100-1800) | 4.63460192176e-07 |
| Smc3_(ab9263) | 4.73160947139e-07 |
| Ctcf | 1.07840967938e-06 |
| Mafk_(ab50322) | 1.44580499132e-06 |
| Corest_(sc-30189) | 1.46790572467e-06 |
| Znf384_(hpa004051) | 2.49911066798e-06 |
| Pparg | 4.65000633568e-06 |
| Gata1_(sc-265) | 8.96286901186e-06 |
| P300 | 1.12819156041e-05 |
| Myod_(sc-32758) | 1.18128120943e-05 |
| Pol2(phosphos2) | 2.55124027888e-05 |
| Usf2 | 2.8128324614e-05 |
| P300_(sc-584) | 5.45813187759e-05 |
| Tal1_(sc-12984) | 0.000167176360024 |
| Tcf12 | 0.000191835667958 |
| Cebpb | 0.000269969087886 |
| Maz_(ab85725) | 0.000293360721942 |
| Srebf1 | 0.000770434432756 |
| Fosl1_(sc-605) | 0.000810462342664 |
| TF | P-Value |
| Pol2-4h8 | 2.28876431892e-09 |
| Pol2(phosphos2) | 4.88289763881e-08 |
| Pol2 | 7.80431537736e-08 |
| Cebpb | 8.11804589727e-08 |
| Srebf1 | 8.35282229172e-07 |
| Ctcf_(sc-15914) | 5.08518495737e-06 |
| Rad21 | 1.54718131929e-05 |
| Pparg | 4.07197746868e-05 |
| Ctcf | 4.38133949259e-05 |
| Gata-1 | 4.60530057163e-05 |
| Myod_(sc-32758) | 5.60439561948e-05 |
| Ets1 | 0.00108865678122 |
| Smc3_(ab9263) | 0.00166563518743 |
| Zkscan1_(hpa006672) | 0.00195750539078 |
| Corest_(sc-30189) | 0.00231520149392 |
| Cry2 | 0.00240738827285 |
| P300 | 0.00262031238724 |
| Myogenin_(sc-12732) | 0.0035498114268 |
| Gata2_(sc-9008) | 0.00532858171868 |
| Mafk_(ab50322) | 0.00536402984575 |
| P300_(sc-584) | 0.00604669894425 |
| Gata1_(sc-265) | 0.00640679132338 |
| Sin3a_(nb600-1263) | 0.00738814553659 |
| E2f4 | 0.00802946144926 |
| Hnf4a | 0.00810634829393 |
| TF | P-Value |
| Pol2(phosphos2) | 3.90422555811e-05 |
| Pol2 | 7.88156429351e-05 |
| Myod_(sc-32758) | 0.000133928070771 |
| P300 | 0.000279117495504 |
| Myogenin_(sc-12732) | 0.00274776608623 |
| Sin3a_(nb600-1263) | 0.00350930305667 |
| Pparg | 0.00482184728891 |
| Ets1 | 0.0050862655946 |
| Gata1_(sc-265) | 0.00525511357511 |
| Hnf4a | 0.00539432175654 |
| Usf2 | 0.0114940287036 |
| Pol2-4h8 | 0.0136875011606 |
| Chd2_(ab68301) | 0.0154266075129 |
| C-myc | 0.0159848246277 |
| Gata-1 | 0.0179157852734 |
| Maz_(ab85725) | 0.025423440935 |
| Ctcf | 0.0270941213523 |
| Tal1_(sc-12984) | 0.0283864896915 |
| Nelfe | 0.0398082903801 |
| Cry1 | 0.0428794514138 |
| Bhlhe40_(nb100-1800) | 0.0570761322261 |
| Nrsf | 0.0621276226274 |
| Corest_(sc-30189) | 0.0675991277822 |
| Cry2 | 0.112964859834 |
| Mxi1_(af4185) | 0.116350713627 |
| TF | P-Value |
| Hnf4a | 1.49741510205e-12 |
| Srebf2 | 2.08150060048e-12 |
| Gata1_(sc-265) | 3.38426215247e-06 |
| Pparg | 2.90177475109e-05 |
| Pol2-4h8 | 5.77520215238e-05 |
| Cbp | 0.000144186965157 |
| Gata-1 | 0.000167592528878 |
| Pol2 | 0.000193790849982 |
| Hcfc1_(nb100-68209) | 0.000245135742979 |
| Nrf2 | 0.000444825103174 |
| C-jun | 0.00051439746782 |
| Pol2(phosphos2) | 0.000615021180153 |
| P300 | 0.00082827216003 |
| Cry1 | 0.000925185977145 |
| P500 | 0.000944749556651 |
| Srebf1 | 0.000958365863035 |
| Gata2_(sc-9008) | 0.00134408858118 |
| Clock | 0.0022458446013 |
| Usf2 | 0.00332754717592 |
| Ctcf | 0.00356135662459 |
| Cry2 | 0.0046029862732 |
| Mafk_(ab50322) | 0.00549821490415 |
| Bhlhe40_(nb100-1800) | 0.009932331645 |
| Rad21 | 0.010077224459 |
| Gcn5 | 0.0111567640305 |
| TF | P-Value |
| Srebf2 | 7.22455215853e-21 |
| Hnf4a | 8.17417438999e-15 |
| Pol2-4h8 | 1.2509604833e-07 |
| Pparg | 7.906671822e-07 |
| Pol2(phosphos2) | 1.7873814517e-06 |
| Mafk_(ab50322) | 8.11076650816e-06 |
| P300_(sc-584) | 1.06507765355e-05 |
| Srebf1 | 2.8887251359e-05 |
| Pol2 | 4.04890375425e-05 |
| Gata1_(sc-265) | 7.59293317751e-05 |
| Nrsf | 0.000508437069615 |
| Ctcf | 0.000801128596029 |
| C-jun | 0.000832926260728 |
| P500 | 0.000893362097675 |
| Usf2 | 0.00115854428995 |
| Gata2_(sc-9008) | 0.00123359398361 |
| Rad21 | 0.00244358055236 |
| E2f4 | 0.00348760025718 |
| Tbp | 0.00403009016165 |
| Smc3_(ab9263) | 0.00516101360987 |
| Gata-1 | 0.00633796003108 |
| Tal1_(sc-12984) | 0.00806389611631 |
| Jund | 0.00998227287651 |
| P300 | 0.0122764087804 |
| Ubf_(sc-13125) | 0.0134694109345 |
| TF | P-Value |
| Hnf4 | 4.03784665477e-40 |
| Ap-2rep | 2.16467752539e-15 |
| Ets2 | 9.69650002769e-15 |
| Pur1 | 7.00588049557e-11 |
| Myogenin | 2.03055006396e-10 |
| Ar | 2.15137144714e-10 |
| Pparg | 2.4183308942e-08 |
| Heb | 1.2592400211e-07 |
| Math1 | 1.02307044332e-06 |
| Lbp9 (tcfcp211) | 4.11213752875e-06 |
| Stat6 | 4.35765714227e-05 |
| Mycn | 0.000118792265833 |
| Mafa | 0.000123424436322 |
| Clock:bmal | 0.000143432030876 |
| Nf-1 | 0.0003867795882 |
| Smad3 | 0.000423523485983 |
| Ctcf | 0.000559960410254 |
| Myb | 0.000671439849312 |
| Hnf4, coup | 0.000761045859304 |
| Myc | 0.00176645686552 |
| Stat3 | 0.00197494816221 |
| Obox5 | 0.00197703951784 |
| C-myb | 0.00223955010518 |
| Pbx | 0.00407352936638 |
| Atf1 | 0.00601162765457 |
| TF | P-Value |
| Srebf2 | 9.49121297193e-20 |
| Hnf4a | 1.67941817494e-07 |
| Srebf1 | 4.23825766962e-07 |
| Nrsf | 9.39728321778e-06 |
| Pol2 | 1.54410483673e-05 |
| Ctcf | 5.38155675662e-05 |
| Max | 0.000205171922312 |
| Pol2(phosphos2) | 0.000580211316266 |
| Maz_(ab85725) | 0.00115105211598 |
| Pol2-4h8 | 0.001532769984 |
| Mxi1_(af4185) | 0.00184789456527 |
| C-myc | 0.00222657533551 |
| Tbp | 0.00257958402857 |
| Pparg | 0.00451199852248 |
| Ubf_(sc-13125) | 0.00760870174909 |
| Sin3a_(nb600-1263) | 0.0106722983546 |
| Myogenin_(sc-12732) | 0.0116003501241 |
| P300_(sc-584) | 0.0143544948345 |
| C-jun | 0.0157537194397 |
| Mafk_(ab50322) | 0.0168478944129 |
| Gata2_(sc-9008) | 0.0179850340945 |
| Nelfe | 0.0274842353784 |
| Gata1_(sc-265) | 0.0451930099114 |
| P500 | 0.0471952982921 |
| Tal1_(sc-12984) | 0.0603864699458 |
| TF | P-Value |
| Srebf2 | 9.0302800529e-07 |
| Pol2-4h8 | 0.00160402315218 |
| Ctcf | 0.00201420178498 |
| Pparg | 0.00258439861576 |
| Sin3a_(nb600-1263) | 0.00436444398385 |
| Hnf4a | 0.00439974054148 |
| Nelfe | 0.0110798523538 |
| Tal1_(sc-12984) | 0.0133190235573 |
| Mafk_(ab50322) | 0.0143129912078 |
| Pol2 | 0.0191966716978 |
| Nrsf | 0.0205328836495 |
| Pol2(phosphos2) | 0.0262115959515 |
| Npas2 | 0.0263217269702 |
| Per2 | 0.0263217269702 |
| Max | 0.030046081435 |
| Hcfc1_(nb100-68209) | 0.030658500866 |
| Tbp | 0.0334967765575 |
| Srebp2 | 0.0458106692368 |
| Znf384_(hpa004051) | 0.0637676045455 |
| Myogenin_(sc-12732) | 0.0743763655728 |
| Fli1_(sc-356) | 0.0870583742189 |
| Usf2 | 0.0921050027 |
| Mxi1_(af4185) | 0.0944087760526 |
| Rad21 | 0.116557022071 |
| Ctcf_(sc-15914) | 0.116557022071 |
| TF | P-Value |
| None |
| TF | P-Value |
| None |
| TF | P-Value |
| Srebf2 | 1.85224201738e-29 |
| Hnf4a | 2.37065516619e-27 |
| Pol2(phosphos2) | 4.22472699723e-16 |
| Pol2 | 1.46690306826e-14 |
| Pparg | 1.57269439628e-13 |
| P300_(sc-584) | 1.07482202714e-11 |
| Rad21 | 5.99089055421e-11 |
| Ctcf_(sc-15914) | 3.65649589465e-10 |
| Gata-1 | 7.79182240861e-09 |
| Mafk_(ab50322) | 2.1787066886e-08 |
| Pol2-4h8 | 2.2891383247e-08 |
| Smc3_(ab9263) | 9.64954416577e-08 |
| Ubf_(sc-13125) | 4.18641405019e-07 |
| Ctcf | 5.49353101511e-07 |
| Usf2 | 1.85162888571e-06 |
| Gata1_(sc-265) | 7.37062388572e-06 |
| Corest_(sc-30189) | 1.182043615e-05 |
| Srebf1 | 1.65280763384e-05 |
| Max | 1.95158118359e-05 |
| C-jun | 2.66647683628e-05 |
| P300 | 3.57949518077e-05 |
| Myogenin_(sc-12732) | 7.02781243538e-05 |
| Ets1 | 8.73866898997e-05 |
| Nrsf | 9.39913045772e-05 |
| Gata2_(sc-9008) | 0.000124123884517 |
| TF | P-Value |
| Hnf4a | 6.66934528611e-30 |
| Srebf2 | 1.18027455587e-28 |
| Gata-1 | 1.50620768397e-18 |
| Ctcf_(sc-15914) | 1.1767953255e-14 |
| Rad21 | 4.41698500963e-14 |
| Pparg | 6.86585792185e-14 |
| Pol2(phosphos2) | 4.14815819652e-13 |
| Smc3_(ab9263) | 8.06187474314e-11 |
| Mafk_(ab50322) | 8.14776865142e-11 |
| Ctcf | 8.69659023664e-11 |
| P300_(sc-584) | 2.04226510906e-10 |
| Pol2-4h8 | 3.19390858485e-10 |
| Gata1_(sc-265) | 4.67490814565e-10 |
| Pol2 | 3.34104975831e-09 |
| P300 | 6.34273899269e-09 |
| Tal1_(sc-12984) | 3.12323726893e-08 |
| Usf2 | 5.057485441e-08 |
| Ets1 | 2.7340111487e-07 |
| Corest_(sc-30189) | 6.71156155137e-07 |
| Bhlhe40_(nb100-1800) | 3.32234357282e-06 |
| C-jun | 4.18056610325e-06 |
| Ubf_(sc-13125) | 6.49942697156e-06 |
| Cry1 | 2.13523154202e-05 |
| Srebf1 | 2.58514807055e-05 |
| Jund | 3.4304686443e-05 |
| TF | P-Value |
| Gata-1 | 3.64490051539e-23 |
| Hnf4a | 1.88690634842e-16 |
| Ctcf_(sc-15914) | 1.42647767508e-13 |
| Rad21 | 2.00739927264e-13 |
| Pparg | 4.382197849e-13 |
| P300 | 8.16056892462e-13 |
| Bhlhe40_(nb100-1800) | 4.71472165173e-11 |
| Smc3_(ab9263) | 8.93996607276e-11 |
| Usf2 | 1.41363972889e-10 |
| Ets1 | 1.98089533656e-10 |
| P300_(sc-584) | 8.89769225824e-10 |
| Gata1_(sc-265) | 1.0257827775e-09 |
| Ctcf | 1.94530168999e-09 |
| Mafk_(ab50322) | 2.2217748139e-09 |
| Corest_(sc-30189) | 1.2630410927e-08 |
| Srebf2 | 1.49725307045e-08 |
| Pol2-4h8 | 1.38228049125e-07 |
| Max | 1.57375908461e-07 |
| Tbp | 3.21598021761e-07 |
| Nrf2 | 4.14936410088e-07 |
| Usf-1 | 4.92834527634e-07 |
| Pol2 | 9.73869268182e-07 |
| Hcfc1_(nb100-68209) | 1.00329489048e-06 |
| Tal1_(sc-12984) | 1.36727288246e-06 |
| Nelfe | 2.0363432264e-06 |
| TF | P-Value |
| Hnf4a | 2.39261915882e-06 |
| Pol2(phosphos2) | 3.91442492415e-06 |
| Gata-1 | 2.46945632237e-05 |
| Ctcf_(sc-15914) | 5.03081233861e-05 |
| Pol2 | 0.000328513375255 |
| Pparg | 0.000371599583526 |
| Corest_(sc-30189) | 0.00065862639763 |
| Bhlhe40_(nb100-1800) | 0.000867972105688 |
| Ctcf | 0.000880100872636 |
| Rad21 | 0.00091550263405 |
| Ets1 | 0.00114735151752 |
| P300_(sc-584) | 0.00162271229052 |
| Mafk_(ab50322) | 0.00165667550726 |
| Znf384_(hpa004051) | 0.00210115123335 |
| Cry2 | 0.00275521413519 |
| Tal1_(sc-12984) | 0.00320121681298 |
| P300 | 0.00376607373566 |
| Gata1_(sc-265) | 0.00488630653475 |
| Bmal | 0.00545595544204 |
| Jund | 0.00634352405335 |
| Cry1 | 0.00775379923779 |
| Smc3_(ab9263) | 0.00791909160422 |
| Tbp | 0.00916676370205 |
| Mxi1_(af4185) | 0.0104676437843 |
| Usf2 | 0.0195830055255 |
| TF | P-Value |
| Rad21 | 0.00012863426603 |
| Mafk_(ab50322) | 0.000153351899835 |
| Ctcf_(sc-15914) | 0.000174531439144 |
| Pol2(phosphos2) | 0.00123622094844 |
| Gata-1 | 0.00133354583096 |
| Pol2-4h8 | 0.00240208407929 |
| Hnf4a | 0.00265033548219 |
| Znf384_(hpa004051) | 0.00544872362036 |
| Ctcf | 0.00572341285552 |
| Gata2_(sc-9008) | 0.00935368403991 |
| Bhlhe40_(nb100-1800) | 0.00986573486444 |
| P300_(sc-584) | 0.0125996990704 |
| Cebpb | 0.0198323291031 |
| Pol2 | 0.0206851088021 |
| Tbp | 0.0358248956274 |
| Ets1 | 0.0401263059458 |
| Per1 | 0.0460236617328 |
| Corest_(sc-30189) | 0.0503241346939 |
| Smc3_(ab9263) | 0.0559661506499 |
| Srebf2 | 0.0643920140319 |
| Pax5_(n-15) | 0.0678701629259 |
| Srebp1 | 0.0736883876905 |
| Pparg | 0.0779909981393 |
| Max | 0.085950791515 |
| Tal1_(sc-12984) | 0.0950543443455 |
| TF | P-Value |
| Pol2 | 0.0100493900867 |
| Gata-1 | 0.051635175813 |
| Pol2(phosphos2) | 0.0538605025986 |
| Maz_(ab85725) | 0.0940975762393 |
| Pax5_(n-15) | 0.132275132275 |
| Gata2_(sc-9008) | 0.156493804497 |
| Hcfc1_(nb100-68209) | 0.199315429173 |
| Ctcf | 0.242239311359 |
| Max | 0.2705548556 |
| Ctcf_(sc-15914) | 0.289959276723 |
| Tal1_(sc-12984) | 0.29926558958 |
| Rad21 | 0.313695150359 |
| Sin3a_(nb600-1263) | 0.32527912273 |
| Srebf1 | 0.384298258507 |
| Nrsf | 0.413196719674 |
| Chd2_(ab68301) | 0.433486479163 |
| Hnf4a | 0.469520226229 |
| Myod_(sc-32758) | 0.475675047886 |
| Tbp | 0.48184988281 |
| Myogenin_(sc-12732) | 0.482229954348 |
| Smc3_(ab9263) | 0.482278891385 |
| C-myc | 0.505800065751 |
| Bmal1 | 0.54233140002 |
| Bmal | 0.542822030889 |
| Cry1 | 0.545037886505 |
| TF | P-Value |
| Pol2 | 4.64697488121e-08 |
| Ctcf_(sc-15914) | 1.10584773142e-05 |
| Hnf4a | 2.89692660906e-05 |
| Rad21 | 7.71952096786e-05 |
| Ctcf | 0.000193335487512 |
| Pol2(phosphos2) | 0.000365480188153 |
| Nrsf | 0.000889648006903 |
| Tal1_(sc-12984) | 0.00226587294246 |
| Myogenin_(sc-12732) | 0.00382343476591 |
| Srebf2 | 0.00395356667212 |
| Max | 0.00586238172228 |
| Cry2 | 0.00837846755846 |
| Pol2-4h8 | 0.00845567217547 |
| Cry1 | 0.00885813522361 |
| Smc3_(ab9263) | 0.009065198853 |
| Ubf_(sc-13125) | 0.0123949246465 |
| Zc3h11a_(nb100-74650 | 0.014819187909 |
| Bmal | 0.0191576851909 |
| Hcfc1_(nb100-68209) | 0.0208537778277 |
| P500 | 0.0286837356611 |
| Usf2 | 0.0342638494829 |
| Pax5_(n-15) | 0.0413443735344 |
| Gata1_(sc-265) | 0.0505415162455 |
| Gcn5 | 0.0511764889479 |
| Clock | 0.0664585399024 |
| TF | P-Value |
| Lbp9 (tcfcp211) | 2.0300997256e-09 |
| Tcfcp2l1 | 0.000303906882103 |
| Nf-kappab | 0.0065355247247 |
| Spz1 | 0.00736222788553 |
| Ctcf | 0.0178506320899 |
| Msx-2 | 0.0372898990987 |
| Heb | 0.055793185099 |
| Srebp-1 | 0.0622387882751 |
| Rush-1alpha | 0.0692674793945 |
| Gcnf | 0.0712304384739 |
| Lef1, tcf1 | 0.106807539139 |
| Foxm1 | 0.114318874921 |
| Hnf3a | 0.118050821861 |
| Sox5 | 0.121566897648 |
| Nobox | 0.131504558464 |
| Hbp1 | 0.145696583247 |
| Ets2 | 0.15227349977 |
| Tal1 | 0.156339687215 |
| Ap-1 | 0.171481385163 |
| Arnt | 0.192978816993 |
| Bach2 | 0.201364572872 |
| Nrsf | 0.202207197019 |
| Math1 | 0.212083668578 |
| Ebf | 0.250380045485 |
| E12 | 0.269133312973 |
| TF | P-Value |
| Myogenin_(sc-12732) | 0.000325875459388 |
| Myod_(sc-32758) | 0.00115343476513 |
| Mafk_(ab50322) | 0.00349530510697 |
| Tcf12 | 0.00558902165442 |
| Pol2 | 0.0119381639226 |
| Hcfc1_(nb100-68209) | 0.0156247559469 |
| Tcf3_(sc-349) | 0.0377225336055 |
| Ctcf | 0.0416623728178 |
| Pol2-4h8 | 0.0464433313781 |
| Srebp2 | 0.0602963530911 |
| P300 | 0.0995500078313 |
| C-myb_(sc-7874) | 0.10618911444 |
| E2f4 | 0.112929305791 |
| Cebpb | 0.115781266788 |
| Max | 0.121264396595 |
| Tal1_(sc-12984) | 0.13852543819 |
| Cry1 | 0.146450752756 |
| Cbp | 0.151238982982 |
| Sin3a_(nb600-1263) | 0.158064985934 |
| Nrf2 | 0.163864429383 |
| Gata1_(sc-265) | 0.190128934915 |
| Ets1 | 0.192311951046 |
| Pparg | 0.212733745805 |
| Ubf_(sc-13125) | 0.227918260789 |
| Ctcf_(sc-15914) | 0.238980125658 |
| TF | P-Value |
| None |
| Pdlim1 | Lancl2 | Cyr61 | Sox30 | Hnrnpab | Twist2 | Fos | Prmt8 |
| Cdkn1c | Fosb | Gsx1 | Klf10 | Yaf2 | Junb | Fhl2 | Adcyap1 |
| Sertad1 | Npas4 | Egr4 | Pcbd1 | Mllt11 | Zfp580 | Fst | Ramp3 |
| Ndnl2 | Sp6 | Mier2 | Med9 | Ddx54 | Pim2 | Zfp653 | Zfp771 |
| Yeats2 | Ercc2 | Skap1 | Zfp263 | Prmt5 | Baz1b | Zfp629 | Zfp579 |
| Sfswap | Nat14 | Psmd9 | Nrf1 | Rnf8 | Batf | Rhog | Habp4 |
| Mapk8ip1 | Ehmt1 | Bhlha15 | Zbed3 | Thap11 | Smyd2 | Zfp664 | Zfp580 |
| Zfp740 | Tcea2 | Vps72 | Dnmt3a | Nkx6-2 | Atf5 | Pbx4 | Cby1 |
| Gtf2h3 | Pelp1 | Tnip1 | Arhgef2 | Hnrnpab | Zfhx2 | Sirt7 | Sox13 |
| Gas6 | Litaf | Ncoa5 | Brms1 | Sin3b | Zscan20 | Sp7 | Gps2 |
| Zfp688 | Zfp524 | Zfp821 | H2afy | Zbtb24 | Banp | Emx1 | Eaf2 |
| Dedd | Zfp239 | Pdlim1 | Batf3 | Csnk2a2 | Tfeb | Nelfb | Ablim2 |
| Zfp46 | Uhrf1 | Hdac11 | Cbx6 | Mapk11 | Cenpb | Hdgf | Trp53inp2 |
| Olig1 | Psrc1 | Neurod2 | Esrra | Tbx21 | Mybbp1a | Dusp26 | Zfp784 |
| Med20 | Med22 | Il1a | Dvl1 | Actl6a | Dlx4 | Ccdc101 | Nr2f1 |
| Cnot8 | Dmap1 | Trf | Bcl11a | Neurog2 | Kat2a | Coprs | Pkig |
| Safb | Rfxank | Zfp423 | Gtf2i | Wnt4 | Alkbh4 | Lime1 | Ell |
| Zbtb49 | Ss18 | Preb | Nr1h2 | Ccnt2 | Irx1 | Ttc5 | Sfrp2 |
| Chaf1a | Nck2 | Pcbd1 | Suv39h3 |
| Cand1 | Sp1 | Drd2 | Topors | Dhx36 | Ehf | Ptprk | Zfp712 |
| Hexb | Heyl | Runx1t1 | Mtdh | Dnttip2 | Ctnnd1 | Rfc1 | Gm4944 |
| Foxo1 | Zfp473 | Zfp748 | Gm6592 | Gli3 | Pcgf5 | Rad21 | Pou2f1 |
| Zbtb16 | Slc11a1 | Nr4a3 | Pbx1 | Ciita | Stat5a | Pias1 | Phf8 |
| Sox11 | Rb1 | Lrp6 | Zfp72 | Sertad2 | Ikzf2 | Mef2a | Ddx3x |
| Zfp458 | Nkrf | Mc1r | Bmpr1a | Mecp2 | Sfpq | Zfp281 | Rbm15 |
| Fosl1 | Cdc73 | 2610305d13rik | Nfe2l2 | Sox5 | Ikzf5 | Tlr2 | Trp53inp1 |
| Zfp169 | Hmbox1 | Tfcp2l1 | Ebf1 | Prrx1 | Zfp516 | Xrn2 | Zfp937 |
| 1700049g17rik | Chd1 | Mina | Cep290 | Ets2 | Bcl6b | Kdm4c | Mapk1 |
| Zfp292 | Fzd4 | Zfp110 | Pkn2 | Crebrf | Trib3 | Wnt1 | Zfp800 |
| Zkscan7 | Jmy | Arid2 | Bbs7 | A530054k11rik | Pitx1 | Ikbkg | Zhx3 |
| Nos1 | Zfp597 | Tle4 | Map2k6 | Rora |
| Ccdc101 | Nkx6-2 | Ell | Pdlim1 | Tfeb | Ablim2 | Pbx4 | Zfp688 |
| Zfp653 | Cbx6 | Habp4 | Mapk11 | Sirt7 | Sox13 | Wnt4 | Trp53inp2 |
| Brms1 | Nrf1 | Mybbp1a | Yeats2 | Zfp46 | Ttc5 | Zfp784 | Chaf1a |
| Irx1 | Med22 | Il1a | Zfp740 | Dlx4 |
| Topors | Trib3 | Pou2f1 | Fzd4 | Hexb | Sox5 | Zkscan7 | Zfp597 |
| Tle4 | Mef2a | Nkrf | Xrn2 | Zfp937 | Gm6592 | Mecp2 |
| Rasl11a | Tesc | Nme2 | Tceb2 | Zfp36 | Klf2 | Cyr61 | Nr4a1 |
| Fos | Zfp580 | Btg2 | Dynll1 | Ndufa13 | Pfdn5 | Egr2 | Fosb |
| Egr4 | Mt3 | Lmo1 | Junb | Jun | Jund | Sertad1 | Edf1 |
| Taf10 | Adcyap1 | Hint1 | Npas4 | Zfp787 | Ramp3 |
| Rasl11a | Csrnp1 | Klf2 | Hes7 | Zfp771 | Prmt8 | Btg2 | Pfn1 |
| Dynll1 | Zfp787 | Hint1 | Dusp26 | Sp2 | Junb | Jund | Ptges2 |
| Adcyap1 | Fosb | Crym | Rps14 | Zfp580 | Trnp1 | Nme2 | Zfp36 |
| Ovol2 | Nr4a2 | Nr4a1 | Ddit3 | Ldb2 | Sertad1 | Taf10 | Ino80b |
| 2410016o06rik | Ramp3 | Nfil3 | Ndn | Pdlim1 | Tesc | Txn1 | Tceb2 |
| Fos | Fosl2 | Klf11 | Nedd8 | Olig1 | Mt3 | Jun | Aes |
| Tgfb1i1 | Znhit1 | Npas4 | Snf8 | Phf5a | Nrarp | Phb | Cited2 |
| Cyr61 | Scand1 | Mrpl12 | Pfdn5 | Egr2 | Egr3 | Egr4 | Rasd1 |
| Lmo1 | Prdx5 | Nupr1 | Fhl2 | Ndufa13 | Edf1 | Id3 | Edn1 |
| Park7 | Sox18 | Snapc2 |
| Chaf1a | Cbx6 | Zfp784 |
| Fzd4 |
| Hes5 | Vhl | 2700060e02rik | Acvr1 | Btg2 | Zfp628 | Zfp629 | Foxs1 |
| Mrpl12 | Psmd9 | Junb | Jund | Phb2 | Zfp444 | Adcyap1 | Zfp511 |
| Zbed3 | Thap11 | Akt1 | Nkx6-2 | Nme2 | Zbtb17 | Ing1 | Ldb2 |
| Ddit3 | Med29 | Chtop | Med27 | Jun | Litaf | 2410016o06rik | Rarg |
| Zfp523 | Zfp524 | Ino80b | H2afy | H2afz | Fgf10 | Mad2l2 | Mypop |
| Lmcd1 | Cd40 | Ppp2r5b | Dnajb5 | Csnk2a2 | Tfdp1 | Tfeb | Tfe3 |
| Gsk3a | Pmf1 | Mapk11 | Kcnip3 | Zfp821 | Edf1 | Zfp13 | Ndnl2 |
| Znhit1 | Zfp787 | Zfp513 | Usp21 | Twist2 | Med28 | Med22 | Cenpb |
| Ramp3 | Dvl1 | Meaf6 | Cnot8 | Zfp956 | Trf | Scand1 | Sra1 |
| Snd1 | Pkig | Hr | Hax1 | Bambi | Fosb | Klf14 | Smarcd3 |
| Fbxw7 | Nr1h2 | Id3 | Nr2f1 | Fam58b | Prdm16 | Lyl1 | Ctbp1 |
| Supt4a | Zfp61 | Rela | Prmt8 | Prmt1 | Prmt7 | Prmt5 | Myd88 |
| Pus1 | Ift57 | Etv5 | Ss18 | E2f1 | Rhog | Gmeb2 | Ehmt1 |
| Dnaja3 | Igf2 | Pomc | Fezf2 | Foxq1 | Pbx4 | Abra | Tbx3 |
| Tbx4 | Hyal2 | Ptov1 | Gas6 | Keap1 | Calr | Stk16 | Phb |
| Zfp672 | Pdlim1 | Tesc | Gtf3a | Creb3l1 | Ablim2 | Fos | Ndn |
| Zfp414 | Foxj1 | Snapc2 | Ywhah | Aes | Dusp26 | Npas4 | Npas1 |
| Preb | Phf5a | Khdrbs3 | Brf2 | Dmap1 | Wbscr22 | Zfp647 | Fzd9 |
| Glo1 | Lhx2 | Zfat | Gtf2b | Lbh | Nat14 | Egr2 | Egr3 |
| Egr4 | Yeats2 | Vegfa | Bag1 | Ndufa13 | Esrra | Bmyc | Sin3b |
| Wbp2 | Tnip1 | Hnrnpab | Pim2 | Skap1 | Ehmt2 | Dnajb6 | Dynll1 |
| Psmc5 | Zfp784 | Sp2 | Csrnp2 | Gtf2ird2 | Tead3 | Tead2 | Nrf1 |
| Rnf10 | Sirt2 | Arhgap22 | Habp4 | Mapk8ip1 | Tsc22d1 | Rps14 | Atf5 |
| Atf1 | Cby1 | Brms1 | Foxg1 | Hic1 | Tcf15 | Prr13 | Mbd3 |
| Ybx1 | Arhgef2 | Sox10 | Sox13 | Vps72 | Taf1c | Nkx3-1 | Sertad1 |
| Taf10 | Hmox1 | Emx1 | Eaf2 | Dedd | Nr1i3 | Abt1 | Mcrs1 |
| Ccdc85b | Sox2 | Pcna | Smarcb1 | Casz1 | Mcts2 | Otx1 | Irf2bp1 |
| Csdc2 | Zbtb7b | Cbx6 | 0610009b22rik | Ctnnbip1 | Olig2 | Olig1 | Pknox1 |
| Bhlha9 | Snf8 | Drap1 | Foxf2 | Dmrta2 | Nrarp | Actl6a | Dlx4 |
| Prkd1 | Ptges2 | Coprs | Kat2a | Cdkn1c | Efcab6 | Smyd2 | Alkbh4 |
| Ell | Zbtb42 | Prdx5 | Nupr1 | Loxl2 | Puf60 | Narfl | Zfp593 |
| Map2k1 | Edn1 | Atoh8 | Park7 | Asf1b | Zfp575 | Rnf187 | Ncoa5 |
| Zfp579 | Med8 | Med9 | Ddx54 | Thrsp | Pfn1 | Sox18 | Tcf25 |
| Tceb2 | Pex14 | Sox17 | Usf2 | Zfp580 | Zfp746 | Zfp740 | Eef1d |
| Chmp1a | Rfxap | Kat5 | Scrt2 | Ssbp3 | Pias4 | Zfp668 | Gsx1 |
| Commd7 | Uhrf1 | Dedd2 | Mapre3 | Tcf3 | Apbb1 | Batf3 | Zfp771 |
| Zfp775 | Maf1 | Rhoa | E430018j23rik | Hdac11 | Fhl2 | Trp53inp2 | Ccnd1 |
| Smad6 | Tgfb1i1 | Ube2i | Hopx | Thap7 | Elp5 | Carhsp1 | Nck2 |
| Elof1 | Ccdc101 | Zfp764 | Zfp768 | Bcl11a | Meox1 | Neurog2 | Ecm1 |
| Lmo1 | Lmo4 | Tagln3 | Thra | Ruvbl1 | Pcbd1 | Rxrb | Setd8 |
| Zfp846 | Zfp318 | Rybp | Bcl6 | Cebpz | Zfp712 | Npat | Runx1t1 |
| Mphosph8 | Mkl2 | Ncl | Supt16 | Grem1 | Rfc1 | Zfp148 | Zbed6 |
| Atf7ip | Tpr | Sfpq | Zbtb16 | Sfmbt1 | Lin54 | Mlxip | Hivep2 |
| Hivep3 | Ccar1 | Crebzf | Notch2 | P2ry1 | Ikzf5 | Ino80d | Ikzf2 |
| Supt20 | Zfp455 | Zfp451 | Zfp458 | L3mbtl1 | Satb2 | Satb1 | Ncoa7 |
| Rb1cc1 | Rnf2 | Tcerg1 | Gsk3b | Spen | Phf14 | Zfp866 | Cdk7 |
| Nipbl | Zfp605 | Zfp322a | Bdp1 | Wwp1 | Zfp516 | Xrn2 | Zfml |
| Zfp462 | Sp110 | Cnot1 | Nfib | Atxn7 | Zfp292 | A430033k04rik | Cpeb3 |
| Zfp189 | Klf12 | Elf1 | Bhlhe40 | Bhlhe41 | Arid2 | Bbs7 | Fzd4 |
| Insr | Ago2 | Id4 | Zfp619 | Chaf1b | Hmg20a | Tenm1 | Tgs1 |
| Dnttip2 | Creb1 | Hlf | Zfp882 | Ppp1r12a | Elk4 | Gli3 | Rad21 |
| Rsl1 | Mga | Tbp | Gabpb2 | Clock | Eif2ak2 | Phf8 | Rb1 |
| Bbx | Bmi1 | Camta1 | Tfcp2l1 | Prdm2 | Pnrc2 | Tardbp | Zzz3 |
| Zfp418 | Onecut2 | Nedd4 | Prkdc | Ezh3 | Hmbox1 | Ralgapa1 | Esrrg |
| Prrx1 | Aire | Chd1 | Chd2 | Chd6 | Chd8 | Eya2 | Ahctf1 |
| Trip4 | Fzd1 | Zfp955b | Jmy | Zfp334 | Ddx17 | Nos1 | Nck1 |
| Vldlr | Zfpm2 | Med13l | Mynn | Cdc5l | Zfp652 | Wwox | Whsc1l1 |
| Nlrc5 | Mtdh | Ago1 | Cir1 | Zfp329 | Zfp326 | Zfp324 | Foxd2 |
| Iws1 | Adrb2 | Gm6592 | Pcgf5 | Khdrbs2 | Atf7 | Has3 | Tgfbr1 |
| Med12l | Nr2c2 | Nr2c1 | Sox11 | Phf21a | Taf1d | Lrp6 | Zfp72 |
| Pik3r1 | Mef2a | Arid4a | Arid4b | N4bp2l2 | Isx | Ash3l | Mecp2 |
| Aff4 | Zfp260 | Hoxd8 | Zfp128 | Sox5 | Bcl11b | Hipk2 | Hipk1 |
| Lpin2 | Hinfp | Map3k9 | Zfp354c | Cdc73 | Whsc1 | Wac | Crebrf |
| Rora | Il4 | Ctnnd1 | Rbl2 | Tfcp2 | Ddx3x | Son | Nrip1 |
| Arid5b | Cav1 | Abcg1 | Kdm4c | Mapk1 | Zfp110 | Rcor1 | A530054k11rik |
| Rps6ka5 | Scai | Osm | Mysm1 | Zfp800 | Zkscan1 | Zkscan2 | Zkscan7 |
| Pitx1 | Zfp599 | Zfp597 | Prkaa2 | Rsf1 | Map2k6 | Ar | Psip1 |
| Cand1 | Kpna6 | Kdm5a | Kdm5b | Kdm5c | Sp1 | Usp47 | Epc2 |
| Csrnp3 | Topors | Rbmx | Dhx33 | Dhx36 | Hexb | Zfp109 | Brca2 |
| Ctr9 | Mycbp2 | Ahr | Zfp960 | Bnc2 | Ccnt1 | Zkscan16 | Setd1b |
| Pygo1 | Trak2 | Erbb4 | Zfp568 | Pias1 | Zfp664 | Jmjd1c | Kdm2a |
| Pdpk1 | Pdcd4 | Lrpprc | Ino80 | Tcf4 | Ascc3 | Nkrf | Tet1 |
| Tet3 | Pou2f1 | Zfp397 | Kdm3a | Nfe2l2 | Hmgn5 | Trp53inp1 | Zfp169 |
| Smad4 | Sfrp1 | Elp4 | Acvr2b | Brd8 | Zbtb33 | Cep290 | Ets2 |
| Phf20 | Zfp760 | T | Epcam | Taf4b | Zfp382 | Tnks | Atrx |
| Ikbkg | Lcor | Zhx3 | Zhx1 | Tef | Ern1 | Gatad2b | Ikbkap |
| Setd7 |
| Zfp579 | Hes5 | Vhl | Gtf3a | Ddx54 | Tnip1 | Hnrnpab | Pim2 |
| Habp4 | Prmt8 | Prmt1 | Prmt5 | Bmyc | Hax1 | Zfp784 | Mapk8ip1 |
| Zfp524 | Pus1 | Pex14 | Nat14 | Etv5 | Nrf1 | Prdm16 | Adcyap1 |
| Gmeb2 | Ehmt1 | Pfn1 | Tsc22d1 | Zfp580 | Stk16 | Zbed3 | Khdrbs3 |
| Chmp1a | Nkx6-2 | Atf5 | Pbx4 | Cby1 | Ddit3 | Tfeb | Hic1 |
| Rps14 | Mbd3 | Ss18 | Sox13 | Taf1c | Zbtb7b | Litaf | Nkx3-1 |
| Sertad1 | 2410016o06rik | Calr | H2afy | Ramp3 | Emx1 | Eaf2 | Actl6a |
| Mad2l2 | Mypop | Ing1 | Pdlim1 | Tesc | Zfp771 | Zfp775 | Tceb2 |
| Mcrs1 | Rhog | Ccdc85b | Ablim2 | Smarcb1 | Casz1 | Otx1 | Kcnip3 |
| Nupr1 | Usf2 | Hdac11 | Cbx6 | Mapk11 | Trp53inp2 | Olig1 | Zfp821 |
| Ccnd1 | Pknox1 | Cdkn1c | Wbp2 | Esrra | Psmd9 | Ndnl2 | Zfp787 |
| Tgfb1i1 | Dusp26 | Hopx | Thap7 | Npas4 | Snf8 | E430018j23rik | Ywhah |
| Elp5 | Med22 | Elof1 | Olig2 | Nrarp | Dlx4 | Bhlha9 | Zfp764 |
| Wbscr22 | Scand1 | Gsx1 | Ptges2 | Coprs | Glo1 | Pkig | Thap11 |
| Sin3b | Bambi | Egr3 | Smyd2 | Alkbh4 | Ell | Jun | Ecm1 |
| Lmo1 | Lmo4 | Preb | Nr1h2 | Puf60 | Fhl2 | Narfl | Edf1 |
| Id3 | Nck2 | Nr2f1 | Park7 | Zfp13 | Rxrb | Setd8 | Ncoa5 |
| Cand1 | Med13l | Kdm5b | Epc2 | Topors | Mycbp2 | Runx1t1 | Mphosph8 |
| Bnc2 | Mkl2 | Ago1 | Zfp326 | Hlf | Rfc1 | Zbed6 | Elk4 |
| Iws1 | Gm6592 | Trak2 | Zfp318 | Phf8 | Son | Hivep3 | Kdm2a |
| Pdcd4 | Taf1d | Lrp6 | Zfp72 | Chd8 | Supt20 | Mef2a | Arid4a |
| Arid4b | N4bp2l2 | Sfrp1 | Mecp2 | Satb1 | Tet1 | Prdm2 | Pou2f1 |
| Erbb4 | Tcerg1 | Zfp397 | Onecut2 | Prkdc | Zfp169 | Hmbox1 | Nipbl |
| Zfp605 | Zfp322a | Bdp1 | Ctnnd1 | Zfp866 | Xrn2 | Ddx3x | Zfp462 |
| Chd2 | Chd6 | Cep290 | Ets2 | Ahctf1 | Atxn7 | Zfp760 | Kdm4c |
| Fzd1 | Zfp955b | Rps6ka5 | Osm | Zkscan2 | Jmy | Lrpprc | Bbs7 |
| Fzd4 | A530054k11rik | Ikbkg | Zhx3 | Mlxip | Aff4 | Id4 | Zfp597 |
| Rsf1 | Map2k6 | Supt16 | Rora | Zfpm2 | Ar | Ikbkap |
| Pdlim1 | Atf5 | Zfp764 | Cby1 | Ddx54 | Pkig | Hdac11 | Cbx6 |
| Mapk11 | Adcyap1 | Trp53inp2 | Olig1 | Pknox1 | Nrf1 | Setd8 | Ehmt1 |
| Calr | Elp5 | Zfp580 |
| Ctnnd1 | Jmy | Bbs7 | Elk4 | Zfp169 | Xrn2 | Supt16 | Son |
| Mlxip |
| Klf4 | Ptrf | Zfp772 | Hnf1b | Kank2 | Foxs1 | E2f7 | Dbx2 |
| Ehmt1 | Zbed3 | Paf1 | Nfkb2 | Irf1 | Zfp36 | Dll4 | Sox17 |
| Rarg | Srebf1 | Tcf7 | Zfp560 | Eaf2 | Mc1r | Plk1 | Ikbkb |
| Prdm1 | Zfp775 | Zbtb7b | Hif3a | Pmf1 | Gata2 | Tspyl2 | Acvrl1 |
| Mybbp1a | Crtc2 | Brd9 | Foxf2 | Zbtb32 | Tfap2e | Mta2 | Lhx3 |
| Park2 | 9130023h24rik | Ell | Pitx1 | Batf2 | Batf3 | Ccnl2 | Foxl2 |
| Id3 | Adnp2 | Prox2 |
| Cand1 | Kdm5b | Sp8 | Epc2 | Zfp719 | Tshz1 | Ntf3 | Lif |
| Rfc1 | Usp22 | Zfp661 | Trak2 | Pcgf5 | Skil | Brms1l | Pdcd4 |
| Hdac2 | Gzf1 | Btrc | Pik3r1 | Mef2c | Mef2a | Ascc3 | Eomes |
| Mecp2 | Gm14124 | Myrfl | Apbb2 | Plcb1 | Fbxw11 | Lpin2 | Khdrbs1 |
| Arnt2 | Neurod1 | Rorb | Zfp605 | Yy1 | Sla2 | Zbtb33 | Mtf1 |
| Mapk1 | Mapk9 | Foxp1 | Zfp189 | Zbtb43 | Jmy | Ppm1a | Sebox |
| Pspc1 | Rgmb | Bmp3 |
| Drd2 | Hes7 | Cebpa | 2700060e02rik | Btg2 | Foxs1 | Inhba | Klf11 |
| Klf16 | Junb | Jund | Adcyap1 | Gsx1 | Pomc | Nme2 | Brf2 |
| Ddit3 | 2410016o06rik | Ino80b | H2afz | Mypop | Pmf1 | Edf1 | Ndnl2 |
| Znhit1 | Twist2 | Mina | Ramp3 | Cited2 | Cyr61 | Scand1 | Snd1 |
| Pkig | Mrpl12 | Fosb | Id2 | Id3 | Rasl11a | Med31 | Ctbp1 |
| Sap30l | Relb | Rela | Prmt8 | Prmt1 | Prmt7 | Hhex | Myd88 |
| Dmrtc1a | Trnp1 | Zfp36 | Stat5a | Abra | Nupr1 | Ldb2 | Bahd1 |
| Calr | Stk16 | Ecd | Phb | Puf60 | Pdlim1 | Tesc | Txn1 |
| Creb3l1 | Fosl2 | Nedd8 | Ndn | Snapc2 | Ywhah | Aes | Dusp26 |
| Npas4 | Phf5a | Erf | Wbscr22 | Fzd9 | Pqbp1 | Lhx2 | Nat14 |
| Egr2 | Egr3 | Egr1 | Egr4 | Rasd1 | Bag1 | Ndufa13 | Esrra |
| Bmyc | Zic1 | Klf4 | Klf2 | Aurkb | Dnajb5 | Gatad1 | Dynll1 |
| Tgfb1i1 | Zfp787 | Rnf187 | Ube2i | Mdk | Mapk8ip1 | Tsc22d1 | Crym |
| Rps14 | Ovol2 | Atf5 | Nr4a2 | Nr4a1 | Cby1 | Prr13 | Mbd3 |
| Ybx1 | Nkx3-1 | Sertad1 | Taf10 | Srebf2 | Sik1 | Tceb2 | Ccdc85b |
| Sox9 | Sox2 | Pcna | Smarcb1 | Mcts2 | Irf2bp2 | Irf2bp1 | Csdc2 |
| Fos | Tlr2 | Olig1 | Mt3 | Churc1 | Hint1 | Snf8 | Drap1 |
| Nrarp | Dlx2 | Sox18 | Ptges2 | Coprs | Heyl | Ptov1 | Prdx5 |
| Map2k1 | Edn1 | Park7 | Sp2 | Csrnp1 | Csrnp2 | Thrsp | Pfn1 |
| Usf2 | Zfp580 | Chmp1a | Pdgfb | Nfil3 | Igf2 | Apbb1 | Zfp771 |
| Dnajc17 | Nelfe | Foxq1 | Mafk | Fhl2 | Eid2 | Srf | Aamdc |
| Ccnd1 | Smad6 | Jun | Thap7 | Elp5 | Elof1 | Zfp764 | Btf3 |
| Atoh7 | Atoh8 | Pfdn5 | Lmo1 | Lmo4 | Tagln3 | Batf2 | Tle4 |
| Pcbd2 | Zfp13 |
| Zbtb7b | Kank2 | 9130023h24rik | Pitx1 | Batf2 | Ccnl2 | Foxl2 | Mc1r |
| Khdrbs1 | Arnt2 | Btrc | Pik3r1 | Gm14124 |
| Foxl2 |
| Arnt2 |
| Ucp1 | Pcgf5 | Csrnp1 | Atf6 | Cyr61 | Nr4a3 | Nr4a1 | Ell2 |
| Fos | Npnt | Fosl2 | Neurog2 | Arid3b | Btg2 | Stat4 | Egr2 |
| Egr3 | Fosb | Egr1 | Egr4 | Inhba | Klf10 | Junb | Zfp36 |
| Sertad1 | Adcyap1 | Npas4 | Gatad2b | Maml3 | Fst |
| Zfp677 | Nr2f2 | Crym | Bcl3 | Mapk3 | Dmrt2 | Dmrt3 | Pmf1 |
| Lmo1 | Tshz2 | Lmo2 | Id3 | Cidea | Mdk | Zfp688 | Pf4 |
| Tle2 | Tcea3 |
| Fos | Npas4 | Btg2 | Junb | Cyr61 | Nr4a3 | Nr4a1 | Egr2 |
| Egr3 | Fosb | Egr1 | Egr4 | Inhba |
| Lmo1 | Cidea |
| Nr4a1 | Npas4 | Btg2 | Fos | Fosb | Junb | Inhba |
| None |
| None |
| Pdlim1 | Zfp281 | Msx3 | Smarca5 | Cyr61 | Nr4a3 | Nr4a1 | Med6 |
| Mcts2 | Hipk3 | Skil | Fos | Crem | Fosl2 | Prmt8 | Arid3b |
| Btg2 | Cdc73 | Dnajb5 | Hspa8 | Per2 | Ell2 | Egr2 | Egr3 |
| Fosb | Egr1 | Per1 | Egr4 | Inhba | Ift57 | Fbxw7 | Etv1 |
| Klf10 | Etv5 | Junb | Esrrg | Fgf9 | Adcyap1 | Sertad1 | Npas4 |
| Tle4 | Nfil3 | Sik1 | Zfp959 | Fst | Larp7 | Zfp948 |
| Hes7 | Tnip2 | Sap18 | Apex1 | Crym | Elk1 | Jmjd6 | Birc5 |
| Irf8 | Foxo6 | Gtf2h3 | Bhlhe22 | Gtf2h4 | Lum | Aebp1 | Phf1 |
| Sox12 | Nodal | Hdac7 | Rreb1 | Srebf1 | Psrc1 | Ascc1 | Banp |
| Zfp956 | Cc2d1b | Gtf3a | Ablim2 | Cbx2 | Crebl2 | Zscan18 | Aamdc |
| Thap7 | Il11 | Il4 | Il1a | Zfp467 | Mapk3 | Rcor2 | Smarcd2 |
| Msc | Tnni2 | Nos1 | Sebox | Taf8 | Bmp4 |
| Pdlim1 | Cyr61 | Nr4a3 | Nr4a1 | Fos | Fosl2 | Prmt8 | Btg2 |
| Dnajb5 | Egr2 | Egr3 | Fosb | Egr1 | Per1 | Egr4 | Inhba |
| Klf10 | Etv5 | Junb | Adcyap1 | Npas4 | Sik1 | Zfp959 | Zfp948 |
| Mapk3 | Smarcd2 | Psrc1 | Apex1 | Foxo6 | Zfp467 |
| Junb | Fosl2 | Etv5 | Fos | Npas4 | Cyr61 | Nr4a3 | Nr4a1 |
| Egr2 | Egr3 | Fosb | Egr1 | Adcyap1 |
| None |
| Six2 | Tcf15 | Mapk12 | Col1a1 | Batf | Ascl3 |
| Prdm12 | Hoxd8 | Hey2 | Nr1h3 | T |
| None |
| None |
| None |
| None |
| Pdlim1 | Cdkn1c | Fst |
| Mitf | Cebpb | Cebpa | Ubp1 | Btg2 | Wasl | Sorbs3 | Tshz1 |
| Hsf1 | Zfhx2 | Junb | Jund | Hes3 | Foxo3 | Foxo4 | Usp2 |
| Nr1d2 | Nkx6-2 | Irf2 | Irf1 | Irf7 | Irf5 | Irf8 | Ppargc1b |
| Zbtb17 | Zbtb16 | Cebpg | Ddit3 | Cebpd | Zfp703 | Crebzf | Brms1 |
| Hcls1 | Litaf | C030039l03rik | Gzf1 | Ptch1 | Epas1 | Nacc2 | Sec14l2 |
| Zfp524 | H2afz | Zfp459 | Mypop | Mepce | Tfeb | Dkk3 | Rnf6 |
| Klf16 | Hace1 | Mapk14 | Mapk11 | Mapk12 | Tspyl2 | Gpbp1l1 | Cdk9 |
| Kcnip3 | Mkx | Zfp14 | Mxi1 | Atmin | Med24 | Brwd1 | Uchl5 |
| Calcoco1 | Cited2 | Cited4 | Nfic | Cyr61 | Trf | Atxn7 | Cpeb3 |
| Klf13 | Ywhah | Hax1 | Klf10 | Klf15 | Bhlhe40 | Bhlhe41 | Fosb |
| Grhl1 | Grhl3 | Mkl2 | Fbxw7 | Nr1h3 | Pkig | Cbfa2t3 | Prickle1 |
| Zfp612 | Id4 | Cc2d1b | Rasl11a | Suds3 | Supt4a | Tet2 | Fgf9 |
| Fgf1 | Sap30l | Prkcb | Etv3 | Etv5 | Ogt | Tcea2 | E2f6 |
| Taf8 | Hlf | Wnt1 | Elk1 | Ppp1r12a | Elk4 | Zfp664 | Dmrtc1a |
| Jdp2 | Shh | Pbx2 | Stat5b | Thrb | Abra | Per3 | Per2 |
| Per1 | Kctd1 | Adrb2 | Dbp | Purb | Bbx | Mkl1 | Camta2 |
| Tfcp2l1 | Vdr | Capn3 | Cry2 | Grin1 | Mlip | Zxdb | Bcl6 |
| Pdlim1 | Tesc | Creb3l3 | Rbm15 | Ablim2 | Gata2 | Fos | Crem |
| Ring1 | Tbk1 | Nab2 | Eya3 | Ezh1 | Acvrl1 | Ndp | Ovol2 |
| Esrrg | Esrra | Bcas3 | Dusp22 | Npas4 | Nr6a1 | Sla2 | Ing5 |
| Ing2 | Pir | Zfp791 | Nat14 | 2410016o06rik | Egr2 | Egr3 | Egr1 |
| Egr4 | Rasd1 | Ndufa13 | Nfkbiz | Rbbp5 | Klf4 | Klf2 | Wbp2 |
| Aebp2 | Tnip2 | Ell3 | Ell2 | Pim2 | Dnajb5 | Xrcc5 | Tceal5 |
| Psmc5 | Trerf1 | Zscan25 | Npas2 | Habp4 | Tsc22d3 | Dyrk1b | Zfp131 |
| Pcgf1 | Pcgf5 | Smarca2 | Atf6 | Slc11a1 | Nr4a1 | Gm14420 | Erf |
| Hbb-y | Il33 | Prr13 | Mbd2 | Arhgef2 | En2 | Hexim1 | Sirt2 |
| Sox10 | Mical2 | Sertad1 | Sema4d | Nrg1 | Frzb | Pou6f1 | Zfp239 |
| Taf11 | Abt1 | Ccdc85b | Sox8 | Sox9 | Irf2bp2 | Hipk1 | Fnip1 |
| Zbtb7a | Zbtb7b | Cbx8 | Cbx7 | Cbx6 | Phf11d | Tlr9 | St18 |
| Mt3 | Psrc1 | Crebl2 | Crebrf | Rorc | Zbtb1 | Rora | Myrf |
| S1pr1 | Ciao1 | Crtc2 | Hsf4 | Zfp691 | Anp32a | Fst | Tsc22d4 |
| Nrip2 | Arid5a | Abcg1 | Atad2 | Mapk1 | Trim66 | Rps6ka4 | Lmcd1 |
| Mlxipl | Gtf2ird2 | Zfp800 | Nupr1 | Atoh7 | Narfl | Wnt10b | Fgfr2 |
| Snw1 | Map2k1 | Edn1 | Il23a | Park2 | Ar | Prox2 | Kdm5a |
| Otud7b | Sp2 | Csrnp1 | Sp7 | Tbr1 | Thrsp | Hexb | Pparg |
| Efna1 | Ppara | Baz1b | Pex14 | Bcor | Zkscan14 | Zkscan16 | Atp8b1 |
| Tcf25 | Zfp367 | Arnt | Pias1 | Hipk3 | Pdcd4 | Nlk | Tbl1xr1 |
| Mapre3 | Zfp827 | Pam | Irak1 | Trnp1 | Rhog | Esrrb | Ccnl1 |
| Creg1 | Mafk | E430018j23rik | Parp14 | Hdac11 | Kdm3a | Srf | Trim30a |
| Smad9 | Smad7 | Jun | Zfp36 | Smad3 | Plag1 | Tlr3 | Adar |
| Cdk7 | Ets2 | Ptpn14 | Actr8 | Runx2 | Neurog2 | Mamld1 | Rreb1 |
| Scx | Ppp3ca | Gabpb2 | Lmo4 | Tagln3 | Ppm1f | Gclc | Batf2 |
| Tef | Fhl2 | Zfpm1 | Ccpg1 | Bmp3 | Rxra | Setd7 | Setd3 |
| Zfp846 | Rybp | Drd2 | Wnt11 | Bcl9 | Hes5 | Hes6 | Phf6 |
| Ahrr | 2700060e02rik | Ccar1 | Mllt1 | Mllt3 | Ercc2 | Ercc3 | Zbtb41 |
| Rnasel | Tshz3 | Foxs1 | Sall3 | Runx1t1 | Wtip | Klf11 | Kdm2b |
| Ncl | Phb2 | Brip1 | Med30 | Adcyap1 | Zfp512 | Jarid2 | Foxo6 |
| Atf7ip | Tpr | Kdm1a | Kdm1b | Akt1 | Uhrf1 | Zeb2 | Top2a |
| Nme2 | Sfpq | Zbtb18 | Zfp521 | Bhlhe22 | Zfpm2 | Gtf2h5 | 3300002i08rik |
| Wdr5 | Gtf2f2 | Rbmxl1 | Flna | Hivep2 | Hivep3 | Id3 | Pou3f2 |
| Pou3f1 | Myt1 | Hdac2 | Hdac6 | Notch2 | Notch3 | Rara | Dnmt3b |
| Ikzf4 | Hmgb1 | Supt20 | Nr2f1 | Vps72 | Mad2l2 | Actl6b | Ilf3 |
| Nhlh2 | L3mbtl1 | L3mbtl3 | Satb1 | Ppp2r5d | Ppp2r5b | Dmbx1 | Ncoa5 |
| Csnk2a1 | Zfp282 | Dkk1 | Rnf4 | Dach2 | Rnf2 | Vopp1 | Zc3h8 |
| F2r | Ctnnd2 | Adora2a | Deaf1 | Gsk3b | Dynll1 | Phf14 | Gtf2e1 |
| Gtf2e2 | Eapp | Phf19 | Tcfl5 | Cdk2 | Cdk8 | Tceal7 | Zfp821 |
| Taf9b | Mxd3 | Suv39h2 | Pid1 | Neurod1 | Neurod6 | Ebf2 | Zfp605 |
| Zfp322a | Tshz2 | Murc | Usp22 | Xrn2 | Cenpf | Sall2 | Ednrb |
| Zfp462 | Rqcd1 | Mapk3 | Hmgb2 | Dvl3 | Pogk | Meaf6 | Cnot1 |
| Emx1 | Gal | Mta2 | Ttf2 | Nfix | Arx | Snd1 | Mycn |
| Pkia | Igsf1 | Kat2a | Rcor1 | Hyal2 | Mrpl12 | Psmd9 | Smarcd3 |
| Phf10 | Arid2 | Ntn3 | Nfatc2ip | Nfxl1 | Pf4 | Irx1 | Zfp28 |
| Chaf1b | Wwc1 | Nr2f2 | Tenm2 | Cc2d1a | Ctbp2 | Tet1 | Dap |
| Cic | Col1a1 | Gtf3a | Ptprk | Ret | Prmt1 | Six1 | Prmt5 |
| Cdkn1c | Map3k9 | Basp1 | Zbtb5 | Rfx3 | Calca | Etv6 | Creb3l1 |
| Smarce1 | E2f7 | Rhoa | Batf | Ehmt1 | Med19 | Zfp229 | Atf7ip2 |
| Zfp146 | Bcorl1 | Tbx1 | Zcchc12 | Trim33 | Igf2 | Cdh13 | Dnmt3a |
| Foxo1 | Nfya | Cdk5rap2 | Pbx4 | Dpf3 | Pbx1 | Zkscan5 | Mmp14 |
| Foxk2 | Zfp866 | Zfp318 | Triap1 | Tbx4 | Lefty1 | Ss18l1 | Gas6 |
| Rb1 | Ldb2 | Pik3r1 | Camta1 | Phtf2 | Dpf1 | Dab2 | Phb |
| Cdh1 | Hif3a | Plk1 | Nfib | Zfp532 | Trp53bp1 | Nfia | Txn1 |
| Prdm8 | Tgif2 | Ablim1 | Ablim3 | Akna | Zfp41 | Zfp40 | Zfp46 |
| Nedd8 | Rbm14 | Ndn | Jmjd1c | Nedd4 | Zfp319 | Snapc3 | Ywhaq |
| Med10 | Ilf2 | Bcl9l | Ezh2 | Chaf1a | Lcorl | Npas1 | Phf5a |
| Trp53inp2 | Med11 | Khdrbs1 | Nr4a3 | C1d | Zfp697 | Chek2 | Tal1 |
| Hnrnpk | Bcl6b | Hnrnpd | Tenm1 | Gtf2a2 | Map2k6 | Zfp647 | Fzd10 |
| Fzd9 | Park7 | Myc | Myb | Zfp955a | Lhx1 | Lhx2 | Lhx6 |
| Zfp426 | Zfp422 | Lbh | Alx4 | Jup | Rnf141 | Xpo1 | Nfatc4 |
| Prdm4 | Zfp579 | Mlxip | Pspc1 | Rgmb | Meox1 | Vldlr | Jak2 |
| Zic3 | Zic1 | Grm5 | Rbbp8 | 2610008e11rik | Psip1 | Klf7 | Klf3 |
| Gm13157 | Klf8 | Snx6 | Picalm | Ascl1 | Hnrnpab | Zfp658 | Hira |
| Wdr77 | Zfp575 | Srsf10 | Dpy30 | Sp5 | Epc1 | Hif1a | Tead2 |
| Tead1 | Ago1 | Ago2 | Mdk | Zfp326 | Zfp324 | Dact1 | Mapk8ip1 |
| Crym | Kdm6b | Birc5 | Ascc1 | Taf1c | Pcgf3 | Pcgf6 | Khdrbs2 |
| Bend6 | Taf1d | Smarca1 | Smarca4 | Kdm5b | Has3 | Tbl1x | Pa2g4 |
| Chd2 | Hic1 | Trp53 | Aebp1 | Aurkb | Tcf12 | Ybx1 | Ybx3 |
| Nono | Hexim2 | Sox11 | Sox12 | Zbtb6 | Zscan21 | Wwtr1 | Naca |
| Zfp90 | Zfp689 | Mesp1 | Tfap2c | Ankrd49 | Mef2c | Foxg1 | Mllt11 |
| Zfp945 | Zfp266 | Taf10 | Prmt3 | Zfp92 | Ccdc101 | Prmt7 | Zfp128 |
| Tceb1 | Irf2bpl | Mcrs1 | Mybl2 | Sox6 | Sox7 | Sox4 | Sox5 |
| Smarcb1 | Glis2 | Bcl11a | Bcl11b | Csdc2 | Fnip2 | Cbx5 | Zfp354c |
| Cbx2 | Cbx1 | Scmh1 | Ctnnbip1 | 5730507c01rik | Whsc1 | Cdca7 | Hip1 |
| Msx3 | Myef2 | Zscan18 | Crtc3 | Hint1 | Taf7 | Ash2l | Zfp760 |
| Zfp184 | Rbl1 | Ilk | Foxf2 | Nrarp | Dlx1 | Rnf168 | Psen1 |
| Nrip1 | Eomes | Lancl2 | Zfp935 | Ccna2 | Kdm4b | Kdm4a | Mapk7 |
| Ckap2 | Fgf10 | Rcor3 | L3mbtl4 | Dnajc2 | Smarcc1 | Hells | Heyl |
| Cdkn1b | Txnip | Ruvbl2 | Prkd2 | Smyd2 | Smyd3 | Zkscan2 | Zbtb46 |
| Loxl2 | Mstn | Nab1 | Zfp592 | Cd40 | Cnot6 | Maf | Satb2 |
| Cask | E2f8 | Sox18 | Lhx5 | Asf1a | Asf1b | Sp8 | Usp47 |
| Csrnp3 | Zfp57 | Zfp54 | Rbmx | Med4 | Dhx36 | Ddx54 | Cops5 |
| Hnrnpa2b1 | Ctr9 | Rbm15b | Tfec | Zfp511 | Jag1 | Ntf3 | Bach1 |
| Galr2 | Mafa | Zfp786 | Ccnt1 | Dach1 | Setd1b | Trp73 | Yeats4 |
| Nkap | Zfp580 | Hsf2 | Ctnnb1 | Chmp1a | Taf6l | Ssbp2 | Zfp663 |
| Otx1 | Asxl3 | Dll1 | Dll4 | Dmrt2 | Arf4 | Zfp583 | Tnks |
| Zfp174 | Lrpprc | Commd7 | Sfswap | Tsg101 | Tbx18 | Eef1d | Zmynd15 |
| Tcf4 | Tcf7 | Tcf3 | Ascc3 | Ascc2 | Cenpk | Irak1bp1 | Trim28 |
| Pmf1 | Vps36 | Jazf1 | Zfp251 | Tet3 | Apbb2 | Igf1 | Mapk15 |
| Bzw1 | Spdef | Egfr | Pelp1 | Scml4 | Trim27 | Dmrtb1 | Hdac10 |
| Foxc1 | Zfp558 | H2afy2 | Zfp423 | Cidea | Eid2 | Rfxap | Eid1 |
| Afap1l2 | Nfe2l3 | 9030624g23rik | Zmiz1 | Lrp5 | Hmgn3 | Hmgn1 | Pcgf2 |
| Ttc5 | Smad4 | Smad5 | Smad1 | Smad2 | Sfrp2 | Sfrp1 | Casz1 |
| Nup62 | Tgfb1i1 | Elp5 | Elp4 | Elp3 | Brd4 | Brd2 | Brd3 |
| Nck2 | Olig2 | Pprc1 | Strap | Ets1 | Zfp946 | Zfp948 | Suv420h1 |
| Suv420h2 | Cdk5rap3 | Thra | Dot1l | Vps25 | Pbx3 | Cav1 | Btf3 |
| Aatf | Sap130 | Npnt | Foxp1 | Rbfox2 | Foxp4 | Nr0b1 | Slc40a1 |
| Wnt7a | Tmsb4x | Lmo1 | Lmo3 | Asxl1 | Atrx | Bmp7 | Eya2 |
| Zfp157 | Trim24 | Nelfcd | Tle3 | Pcbd1 | Lum | Dnmt1 | Rfx7 |
| Bmp2 | Ikbkap |
| Cebpa | Btg2 | Wasl | Sorbs3 | Zfhx2 | Junb | Foxo4 | Usp2 |
| Nr1d2 | Nkx6-2 | Irf2 | Irf1 | Irf7 | Zbtb17 | Zbtb16 | Ddit3 |
| Brms1 | Epas1 | Sec14l2 | Mypop | Dkk3 | Rnf6 | Hace1 | Mapk14 |
| Mapk11 | Tspyl2 | Gpbp1l1 | Kcnip3 | Mkx | Mxi1 | Atmin | Med24 |
| Calcoco1 | Cited4 | Nfic | Cyr61 | Trf | Cpeb3 | Hax1 | Klf10 |
| Klf15 | Bhlhe40 | Bhlhe41 | Fosb | Grhl1 | Grhl3 | Mkl2 | Nr1h3 |
| Pkig | Prickle1 | Zfp612 | Id4 | Supt4a | Fgf1 | Etv3 | Etv5 |
| Tcea2 | Hlf | Elk1 | Zfp664 | Dmrtc1a | Jdp2 | Shh | Pbx2 |
| Stat5b | Thrb | Per3 | Per2 | Per1 | Kctd1 | Dbp | Bbx |
| Camta2 | Capn3 | Grin1 | Pdlim1 | Tesc | Ablim2 | Fos | Crem |
| Ring1 | Nab2 | Ezh1 | Ovol2 | Esrra | Npas4 | Ing2 | Pir |
| Nat14 | Egr2 | Egr3 | Egr1 | Egr4 | Rasd1 | Wbp2 | Aebp2 |
| Tnip2 | Ell2 | Tceal5 | Trerf1 | Habp4 | Tsc22d3 | Pcgf5 | Smarca2 |
| Atf6 | Nr4a1 | Erf | Il33 | Prr13 | Mbd2 | Hexim1 | Sirt2 |
| Sox10 | Mical2 | Sertad1 | Sema4d | Frzb | Taf11 | Sox8 | Fnip1 |
| Zbtb7a | Zbtb7b | Cbx7 | Cbx6 | St18 | Mt3 | Psrc1 | Crebl2 |
| Crebrf | Rorc | Rora | Myrf | S1pr1 | Hsf4 | Nrip2 | Arid5a |
| Abcg1 | Trim66 | Atoh7 | Wnt10b | Snw1 | Map2k1 | Edn1 | Ar |
| Otud7b | Csrnp1 | Sp7 | Tbr1 | Thrsp | Hexb | Pparg | Baz1b |
| Zkscan16 | Zfp367 | Pdcd4 | Nlk | Tbl1xr1 | Mapre3 | Pam | Trnp1 |
| Rhog | Hdac11 | Kdm3a | Srf | Smad7 | Zfp36 | Smad3 | Plag1 |
| Tlr3 | Adar | Ets2 | Runx2 | Rreb1 | Ppp3ca | Ppm1f | Tef |
| Fhl2 | Zfpm1 | Bmp3 | Rxra | Setd7 | Setd3 |
| Zfp846 | Drd2 | Bcl9 | Hes6 | Phf6 | Mllt1 | Zbtb41 | Rnasel |
| Tshz3 | Runx1t1 | Kdm2b | Foxo6 | Kdm1b | Uhrf1 | Top2a | Nme2 |
| Sfpq | Zbtb18 | Bhlhe22 | Gtf2h5 | Wdr5 | Gtf2f2 | Flna | Hivep3 |
| Pou3f2 | Pou3f1 | Hdac2 | Notch3 | Rara | L3mbtl1 | L3mbtl3 | Ppp2r5b |
| Ncoa5 | Zfp282 | Rnf2 | F2r | Deaf1 | Gsk3b | Phf14 | Gtf2e1 |
| Cdk2 | Tceal7 | Zfp821 | Pid1 | Neurod1 | Neurod6 | Usp22 | Xrn2 |
| Cenpf | Sall2 | Ednrb | Zfp462 | Rqcd1 | Hmgb2 | Gal | Nfix |
| Snd1 | Mycn | Pkia | Igsf1 | Hyal2 | Chaf1b | Wwc1 | Nr2f2 |
| Tenm2 | Ctbp2 | Tet1 | Cic | Col1a1 | Gtf3a | Ptprk | Ret |
| Prmt1 | Prmt5 | Basp1 | Zbtb5 | Rfx3 | Calca | Etv6 | Creb3l1 |
| Smarce1 | E2f7 | Ehmt1 | Zcchc12 | Igf2 | Cdh13 | Dnmt3a | Nfya |
| Mmp14 | Foxk2 | Ldb2 | Phtf2 | Dpf1 | Dab2 | Plk1 | Trp53bp1 |
| Txn1 | Ablim1 | Ablim3 | Zfp41 | Zfp46 | Ndn | Ywhaq | Med10 |
| Ilf2 | Bcl9l | Ezh2 | Nr4a3 | C1d | Bcl6b | Tenm1 | Fzd9 |
| Myc | Zfp955a | Lhx6 | Lbh | Jup | Rnf141 | Pspc1 | Rgmb |
| Vldlr | Klf7 | Klf8 | Picalm | Hnrnpab | Dpy30 | Epc1 | Tead2 |
| Ago1 | Mdk | Zfp324 | Dact1 | Kdm6b | Birc5 | Pcgf3 | Bend6 |
| Has3 | Aurkb | Ybx1 | Ybx3 | Nono | Sox11 | Sox12 | Wwtr1 |
| Zfp90 | Mesp1 | Mllt11 | Mcrs1 | Sox7 | Sox4 | Sox5 | Glis2 |
| Bcl11a | Bcl11b | Csdc2 | Zfp354c | Ctnnbip1 | Whsc1 | Hip1 | Myef2 |
| Ash2l | Ilk | Nrarp | Dlx1 | Nrip1 | Ccna2 | Kdm4b | Kdm4a |
| Mapk7 | Ckap2 | Rcor3 | L3mbtl4 | Txnip | Smyd2 | Loxl2 | Mstn |
| Maf | Satb2 | Cask | Csrnp3 | Zfp57 | Rbmx | Cops5 | Rbm15b |
| Bach1 | Trp73 | Zfp580 | Taf6l | Ssbp2 | Asxl3 | Dll1 | Dll4 |
| Arf4 | Tnks | Sfswap | Tbx18 | Tcf4 | Tcf7 | Irak1bp1 | Vps36 |
| Jazf1 | Tet3 | Igf1 | Mapk15 | Bzw1 | Spdef | Scml4 | Dmrtb1 |
| Hdac10 | Zfp558 | H2afy2 | Eid1 | Afap1l2 | Hmgn3 | Smad1 | Sfrp2 |
| Nup62 | Tgfb1i1 | Elp3 | Brd2 | Brd3 | Ets1 | Zfp946 | Suv420h2 |
| Pbx3 | Cav1 | Btf3 | Npnt | Rbfox2 | Slc40a1 | Tmsb4x | Lmo3 |
| Bmp7 | Zfp157 | Pcbd1 | Ikbkap |
| Mitf | Cebpb | Cebpa | Btg2 | Wasl | Sorbs3 | Zfhx2 | Junb |
| Jund | Hes3 | Foxo3 | Foxo4 | Usp2 | Nr1d2 | Nkx6-2 | Irf2 |
| Irf1 | Irf7 | Irf8 | Zbtb17 | Zbtb16 | Ddit3 | Cebpd | Crebzf |
| Brms1 | Ptch1 | Epas1 | Nacc2 | Sec14l2 | Zfp459 | Mypop | Dkk3 |
| Rnf6 | Klf16 | Hace1 | Mapk14 | Mapk11 | Mapk12 | Tspyl2 | Gpbp1l1 |
| Cdk9 | Kcnip3 | Mkx | Mxi1 | Atmin | Med24 | Calcoco1 | Cited4 |
| Nfic | Cyr61 | Trf | Cpeb3 | Klf13 | Hax1 | Klf10 | Klf15 |
| Bhlhe40 | Bhlhe41 | Fosb | Grhl1 | Grhl3 | Mkl2 | Fbxw7 | Nr1h3 |
| Pkig | Cbfa2t3 | Prickle1 | Zfp612 | Id4 | Cc2d1b | Rasl11a | Supt4a |
| Fgf9 | Fgf1 | Prkcb | Etv3 | Etv5 | Tcea2 | Hlf | Elk1 |
| Ppp1r12a | Zfp664 | Dmrtc1a | Jdp2 | Shh | Pbx2 | Stat5b | Thrb |
| Abra | Per3 | Per2 | Per1 | Kctd1 | Dbp | Bbx | Camta2 |
| Tfcp2l1 | Capn3 | Cry2 | Grin1 | Zxdb | Pdlim1 | Tesc | Rbm15 |
| Ablim2 | Gata2 | Fos | Crem | Ring1 | Tbk1 | Nab2 | Ezh1 |
| Ovol2 | Esrra | Bcas3 | Npas4 | Nr6a1 | Sla2 | Ing2 | Pir |
| Nat14 | Egr2 | Egr3 | Egr1 | Egr4 | Rasd1 | Nfkbiz | Klf2 |
| Wbp2 | Aebp2 | Tnip2 | Ell2 | Pim2 | Dnajb5 | Xrcc5 | Tceal5 |
| Psmc5 | Trerf1 | Habp4 | Tsc22d3 | Dyrk1b | Zfp131 | Pcgf1 | Pcgf5 |
| Smarca2 | Atf6 | Slc11a1 | Nr4a1 | Erf | Hbb-y | Il33 | Prr13 |
| Mbd2 | Hexim1 | Sirt2 | Sox10 | Mical2 | Sertad1 | Sema4d | Nrg1 |
| Frzb | Pou6f1 | Zfp239 | Taf11 | Ccdc85b | Sox8 | Sox9 | Irf2bp2 |
| Hipk1 | Fnip1 | Zbtb7a | Zbtb7b | Cbx8 | Cbx7 | Cbx6 | St18 |
| Mt3 | Psrc1 | Crebl2 | Crebrf | Rorc | Rora | Myrf | S1pr1 |
| Crtc2 | Hsf4 | Fst | Tsc22d4 | Nrip2 | Arid5a | Abcg1 | Trim66 |
| Mlxipl | Atoh7 | Wnt10b | Snw1 | Map2k1 | Edn1 | Il23a | Park2 |
| Ar | Prox2 | Otud7b | Sp2 | Csrnp1 | Sp7 | Tbr1 | Thrsp |
| Hexb | Pparg | Efna1 | Baz1b | Zkscan14 | Zkscan16 | Zfp367 | Hipk3 |
| Pdcd4 | Nlk | Tbl1xr1 | Mapre3 | Zfp827 | Pam | Irak1 | Trnp1 |
| Rhog | Mafk | E430018j23rik | Parp14 | Hdac11 | Kdm3a | Srf | Smad7 |
| Jun | Zfp36 | Smad3 | Plag1 | Tlr3 | Adar | Ets2 | Ptpn14 |
| Actr8 | Runx2 | Neurog2 | Rreb1 | Ppp3ca | Lmo4 | Tagln3 | Ppm1f |
| Gclc | Batf2 | Tef | Fhl2 | Zfpm1 | Ccpg1 | Bmp3 | Rxra |
| Setd7 | Setd3 |
| Zfp846 | Rybp | Drd2 | Bcl9 | Hes6 | Phf6 | 2700060e02rik | Ccar1 |
| Mllt1 | Ercc2 | Ercc3 | Zbtb41 | Rnasel | Tshz3 | Sall3 | Runx1t1 |
| Wtip | Klf11 | Kdm2b | Phb2 | Brip1 | Zfp512 | Foxo6 | Kdm1a |
| Kdm1b | Akt1 | Uhrf1 | Top2a | Nme2 | Sfpq | Zbtb18 | Bhlhe22 |
| Gtf2h5 | Wdr5 | Gtf2f2 | Rbmxl1 | Flna | Hivep2 | Hivep3 | Pou3f2 |
| Pou3f1 | Myt1 | Hdac2 | Notch3 | Rara | Hmgb1 | Supt20 | Nr2f1 |
| Mad2l2 | L3mbtl1 | L3mbtl3 | Satb1 | Ppp2r5d | Ppp2r5b | Ncoa5 | Zfp282 |
| Dach2 | Rnf2 | F2r | Deaf1 | Gsk3b | Phf14 | Gtf2e1 | Gtf2e2 |
| Tcfl5 | Cdk2 | Tceal7 | Zfp821 | Mxd3 | Pid1 | Neurod1 | Neurod6 |
| Zfp605 | Tshz2 | Usp22 | Xrn2 | Cenpf | Sall2 | Ednrb | Zfp462 |
| Rqcd1 | Hmgb2 | Dvl3 | Pogk | Meaf6 | Gal | Nfix | Arx |
| Snd1 | Mycn | Pkia | Igsf1 | Hyal2 | Smarcd3 | Phf10 | Arid2 |
| Zfp28 | Chaf1b | Wwc1 | Nr2f2 | Tenm2 | Cc2d1a | Ctbp2 | Tet1 |
| Cic | Col1a1 | Gtf3a | Ptprk | Ret | Prmt1 | Prmt5 | Cdkn1c |
| Map3k9 | Basp1 | Zbtb5 | Rfx3 | Calca | Etv6 | Creb3l1 | Smarce1 |
| E2f7 | Ehmt1 | Med19 | Zfp229 | Zfp146 | Zcchc12 | Igf2 | Cdh13 |
| Dnmt3a | Nfya | Dpf3 | Pbx1 | Mmp14 | Foxk2 | Triap1 | Rb1 |
| Ldb2 | Phtf2 | Dpf1 | Dab2 | Phb | Hif3a | Plk1 | Nfib |
| Trp53bp1 | Txn1 | Prdm8 | Ablim1 | Ablim3 | Zfp41 | Zfp40 | Zfp46 |
| Ndn | Jmjd1c | Zfp319 | Ywhaq | Med10 | Ilf2 | Bcl9l | Ezh2 |
| Chaf1a | Lcorl | Med11 | Khdrbs1 | Nr4a3 | C1d | Chek2 | Bcl6b |
| Hnrnpd | Tenm1 | Gtf2a2 | Zfp647 | Fzd9 | Myc | Zfp955a | Lhx2 |
| Lhx6 | Zfp426 | Zfp422 | Lbh | Jup | Rnf141 | Nfatc4 | Prdm4 |
| Zfp579 | Mlxip | Pspc1 | Rgmb | Meox1 | Vldlr | Zic3 | Grm5 |
| Psip1 | Klf7 | Gm13157 | Klf8 | Snx6 | Picalm | Ascl1 | Hnrnpab |
| Hira | Wdr77 | Zfp575 | Dpy30 | Sp5 | Epc1 | Hif1a | Tead2 |
| Tead1 | Ago1 | Ago2 | Mdk | Zfp326 | Zfp324 | Dact1 | Mapk8ip1 |
| Crym | Kdm6b | Birc5 | Pcgf3 | Pcgf6 | Khdrbs2 | Bend6 | Taf1d |
| Smarca1 | Smarca4 | Has3 | Tbl1x | Pa2g4 | Aebp1 | Aurkb | Tcf12 |
| Ybx1 | Ybx3 | Nono | Hexim2 | Sox11 | Sox12 | Zbtb6 | Zscan21 |
| Wwtr1 | Zfp90 | Zfp689 | Mesp1 | Mef2c | Foxg1 | Mllt11 | Zfp266 |
| Prmt3 | Ccdc101 | Mcrs1 | Sox6 | Sox7 | Sox4 | Sox5 | Glis2 |
| Bcl11a | Bcl11b | Csdc2 | Cbx5 | Zfp354c | Ctnnbip1 | Whsc1 | Cdca7 |
| Hip1 | Myef2 | Hint1 | Ash2l | Zfp760 | Zfp184 | Rbl1 | Ilk |
| Foxf2 | Nrarp | Dlx1 | Nrip1 | Eomes | Lancl2 | Ccna2 | Kdm4b |
| Kdm4a | Mapk7 | Ckap2 | Rcor3 | L3mbtl4 | Dnajc2 | Smarcc1 | Txnip |
| Smyd2 | Loxl2 | Mstn | Nab1 | Cd40 | Maf | Satb2 | Cask |
| Asf1b | Csrnp3 | Zfp57 | Zfp54 | Rbmx | Med4 | Ddx54 | Cops5 |
| Hnrnpa2b1 | Ctr9 | Rbm15b | Ntf3 | Bach1 | Zfp786 | Trp73 | Nkap |
| Zfp580 | Taf6l | Ssbp2 | Asxl3 | Dll1 | Dll4 | Arf4 | Zfp583 |
| Tnks | Sfswap | Tsg101 | Tbx18 | Eef1d | Tcf4 | Tcf7 | Ascc3 |
| Irak1bp1 | Trim28 | Pmf1 | Vps36 | Jazf1 | Zfp251 | Tet3 | Igf1 |
| Mapk15 | Bzw1 | Spdef | Egfr | Pelp1 | Scml4 | Trim27 | Dmrtb1 |
| Hdac10 | Zfp558 | H2afy2 | Cidea | Eid1 | Afap1l2 | Zmiz1 | Hmgn3 |
| Hmgn1 | Pcgf2 | Smad4 | Smad5 | Smad1 | Smad2 | Sfrp2 | Sfrp1 |
| Nup62 | Tgfb1i1 | Elp4 | Elp3 | Brd2 | Brd3 | Pprc1 | Ets1 |
| Zfp946 | Suv420h2 | Cdk5rap3 | Thra | Vps25 | Pbx3 | Cav1 | Btf3 |
| Sap130 | Npnt | Foxp1 | Rbfox2 | Foxp4 | Nr0b1 | Slc40a1 | Wnt7a |
| Tmsb4x | Lmo1 | Lmo3 | Atrx | Bmp7 | Zfp157 | Trim24 | Nelfcd |
| Tle3 | Pcbd1 | Dnmt1 | Ikbkap |
| Pkd1 | Zfhx2 | Scrt1 | Hes3 | Prmt8 | Cebpb | Cebpa | Phf1 |
| Ubp1 | Ciao1 | Btg2 | Srebf1 | Sorbs3 | Tshz1 | Tbx6 | Junb |
| Jund | Mitf | Lime1 | Zfp276 | Zfp513 | Foxo4 | Thap11 | Cela1 |
| Usp2 | Nr1d1 | Nr1d2 | Nkx6-2 | Irf2 | Irf1 | Irf7 | Irf5 |
| Irf8 | Bcas3 | Zbtb17 | Ddit3 | Gtf2h4 | Gtf2f1 | Zfp703 | Med28 |
| Med27 | Crebzf | Brms1 | Jun | Hdac7 | Hdac8 | Litaf | Epas1 |
| 2410016o06rik | Nacc2 | Sec14l2 | Zfp523 | Zfp524 | Ino80b | H2afz | Hmga1 |
| Yap1 | Mypop | Mepce | Tfeb | Dkk3 | Rnf6 | Klf16 | Tfe3 |
| Hace1 | Agap2 | Mapk14 | Mapk11 | Mapk12 | Tspyl2 | Itga8 | Gpbp1l1 |
| Zfp865 | Cdk5 | Cdk9 | Kcnip3 | Tlr3 | Creb3l2 | Rsc1a1 | Mkx |
| Zfp827 | Neurod4 | Zfp14 | Zfp12 | Ndnl2 | Dlx4 | Mbtps1 | Wasl |
| Zfp784 | Zfp787 | Mxi1 | Twist2 | Atmin | Med20 | Med24 | Ttf1 |
| Uchl5 | Calcoco1 | Cited2 | Cited4 | Nfic | Cyr61 | Trf | Atxn7 |
| Sra1 | Coprs | Pkig | Cpeb3 | Tmem173 | Hax1 | Klf10 | Klf15 |
| Bhlhe40 | Bhlhe41 | Fosb | Wnt4 | Fzd2 | Grhl1 | Grhl3 | Mkl1 |
| Mkl2 | Fbxw7 | Fhl2 | Nr1h2 | Prickle1 | Zkscan6 | Acvrl1 | Zfp612 |
| Id4 | Nr2f6 | Fam58b | Safb | Ctbp1 | Supt4a | Dbx2 | Apbb1 |
| Fgf9 | Fgf1 | Zfp1 | Gtf2i | Sap30l | Dab2ip | Zfp692 | Pus1 |
| Etv3 | Etv5 | Ogt | 9130019o22rik | E2f1 | Gmeb2 | Hlf | Elk1 |
| Zfp488 | Zfp664 | Dmrtc1a | Spata24 | Shh | Pbx2 | Stat5b | Lmcd1 |
| Thrb | Abra | Zbtb49 | Per3 | Per2 | Per1 | Kctd1 | Adrb2 |
| Dbp | Keap1 | Gps2 | Camta2 | Nr1h3 | Birc2 | Capn3 | Mlip |
| Zxdb | Zfp672 | Puf60 | Mamstr | Crlf3 | Pdlim1 | Tesc | Creb3l3 |
| Ablim2 | Gata2 | Crem | Grin1 | Zfp414 | Rbm10 | Ring1 | Pml |
| Hdgf | Tbk1 | Eya3 | Ezh1 | Aes | Esrra | Esrrb | Atf4 |
| Dusp22 | Npas4 | Npas2 | Preb | Sebox | Sla2 | Ywhah | Hmg20b |
| Taf6 | Safb2 | Snw1 | Arap1 | Aebp2 | Bhlha9 | Zfp36 | Ing2 |
| Wbscr22 | Pir | Zfp101 | Trim11 | Nat14 | Egr2 | Egr3 | Egr1 |
| Egr4 | Ncor2 | Atad2 | Rasd1 | Ndufa13 | Nfkbiz | Bmyc | Creg1 |
| Rbbp5 | Cux2 | Klf2 | Wbp2 | Tnip2 | Tnip1 | Ell2 | Pim2 |
| Dvl1 | Skap1 | Dnajb5 | Xrcc5 | Tceal5 | Psmc5 | Sp2 | Lpin1 |
| Hopx | Gtf2ird2 | Zfp775 | Trerf1 | Zscan25 | Habp4 | Sox17 | Tsc22d3 |
| Jdp2 | Dyrk1b | Rps14 | Il1a | Tcea2 | Cry2 | Zfp131 | Pcgf1 |
| Pcgf5 | Atf5 | Smarca2 | Atf6 | Atf1 | Slc11a1 | Nr4a1 | Erf |
| Il33 | Tcf15 | Prr13 | Mbd3 | Mbd2 | Arhgef2 | En2 | Hexim1 |
| Sirt2 | Sirt7 | Sox10 | Sox13 | Mical2 | Tfip11 | Sertad1 | Taf11 |
| Zfp688 | Nrg1 | Psrc1 | Frzb | Eaf2 | Pou6f1 | Zfp235 | Zfp239 |
| Zfp93 | Nacc1 | Sema4d | Ccdc85b | Sox8 | Ptpn14 | Irf2bp2 | Hipk1 |
| Fnip1 | Zbtb7a | Fos | Cbx8 | Cbx7 | Cbx6 | Cbx4 | Flcn |
| Brwd1 | St18 | Olig1 | Crebl2 | Crebrf | Mdm2 | Ctdsp1 | Helz2 |
| Rorc | Rora | H2afy | Myrf | S1pr1 | Crtc1 | Crtc2 | Hsf1 |
| Hsf4 | Rfx1 | Zfp764 | Snf8 | Drap1 | Zfp691 | Tbx22 | Fst |
| Tsc22d4 | Nrip2 | Arid5a | Ddx41 | Abcg1 | Ptges2 | Zfp112 | Trim66 |
| Rps6ka4 | Suds3 | Mlxipl | Ell | Prdx5 | Nupr1 | Dpf2 | Atoh7 |
| Ccnt2 | Narfl | Nab2 | Wnt10b | Taf8 | Map2k1 | Edn1 | Il23a |
| Park2 | Paxip1 | Prox2 | Cbfa2t3 | Zfp574 | Otud7b | Dapk3 | Nog |
| Csrnp1 | Sp7 | Tbr1 | Med9 | Thrsp | Hexb | Pparg | Ppard |
| Abt1 | Baz1b | Pex14 | Zkscan14 | Zkscan16 | Atp8b1 | Efna1 | Tcf25 |
| Ldb1 | Usf2 | Zfp740 | Kat5 | Zfp367 | Arnt | Ssbp3 | Pias4 |
| Pdcd4 | Nlk | Zbtb7b | Ccnl2 | Dedd2 | Tbl1xr1 | Mapre3 | Pam |
| Irak1 | Zfp956 | Cc2d1b | Trnp1 | Tet2 | Batf3 | Tnfrsf4 | Zfp771 |
| Zfp777 | Maf1 | Rhog | Ccnl1 | Nelfe | Supt5 | Mafk | E430018j23rik |
| Parp14 | Hdac11 | Kdm3a | Zbtb22 | Srf | Trim30a | Aamdc | Bbx |
| Smad9 | Smad7 | Smad3 | Thap7 | Plag1 | Adar | Ovol2 | Elof1 |
| Ets2 | Nfkbia | Caprin2 | Mt3 | Pomc | Actr8 | Runx2 | Neurog2 |
| Mamld1 | Rreb1 | Pfdn5 | Ppp3ca | Lmo4 | Tagln3 | Ppm1f | Gclc |
| Tef | Edf1 | Zfpm1 | Rxrb | Pcbd2 | Fhod1 | Ccpg1 | Rxra |
| E2f6 | Setd3 | Setd7 |
| Zfp846 | Ehf | Rybp | Bcl9 | Hes6 | Ahrr | 2700060e02rik | Acvr1 |
| Ccar1 | Zfp712 | Khsrp | Mllt1 | Zscan18 | Hexim2 | Rnasel | Tshz2 |
| Tshz3 | Sall3 | Sall2 | Drd2 | Runx1t1 | Zbtb44 | Klf11 | Ncl |
| Gabpa | Mynn | Phb2 | Supt16 | Brip1 | Cir1 | Cdca7 | Zfp512 |
| Jarid2 | Commd7 | Apex1 | Ss18l1 | Trim33 | Foxo1 | Foxo6 | Atf7ip |
| Tpr | Rlim | Kdm1a | Kdm1b | Uhrf1 | Tceb3 | Top2a | Nme2 |
| Eapp | Ppargc1a | Sfpq | Zbtb14 | Zbtb18 | Zfp521 | Bhlhe22 | Zfpm2 |
| Gtf2h5 | Lin54 | Wdr5 | Gtf2f2 | Rbmxl1 | Flna | Dpf3 | Hivep2 |
| Hivep3 | Cebpz | Pou3f3 | Pou3f2 | Pou3f1 | Pou3f4 | Myt1 | Hdac2 |
| Hdac4 | Hdac9 | Notch2 | Notch3 | Phb | Rara | Zfp719 | Zfp462 |
| Usp13 | Pogk | Rnf4 | Mllt3 | Hmgb2 | Ikzf4 | Hmgb1 | Dmrta1 |
| Supt20 | Brd7 | Zfp451 | Zfp458 | Nr2f1 | Fgf10 | Mad2l2 | Bmi1 |
| Bmpr1a | Nhlh2 | L3mbtl1 | L3mbtl3 | Satb1 | Ppp2r5d | Ppp2r5b | Foxn3 |
| Ercc3 | Dmbx1 | Ncoa5 | Actl6b | Zfp282 | Mapk8ip1 | Nostrin | Dach2 |
| Rnf2 | Vopp1 | Zc3h8 | F2r | Ctnnd2 | Setd2 | Adora2a | Deaf1 |
| Gsk3b | Spen | Cnot6 | Gm13157 | Phf14 | Gtf2e1 | Gtf2e2 | Zfp26 |
| Mospd1 | Hdac10 | Tcfl5 | Cdk2 | Tceal7 | Zfp821 | Tcf7 | Aatf |
| Taf9b | Mxd3 | Suv39h2 | Zfp623 | Pid1 | Neurod1 | Neurod6 | Ebf3 |
| Ebf2 | Zfp605 | Zfp607 | Zfp322a | Zfp72 | Cbfb | Murc | Nfib |
| Cenpk | Usp22 | Dkk1 | Xrn2 | Cenpf | Plk1 | Zfml | Ednrb |
| Rqcd1 | Mapk3 | Dvl3 | Meaf6 | Cnot1 | Cnot7 | Sap130 | Irak1bp1 |
| Cd36 | Gal | Nfia | Zfp292 | Bdp1 | Ttf2 | Nfix | Arx |
| Igsf1 | Snd1 | Pkia | 1700049g17rik | Hyal2 | Klf12 | Smarcd3 | Zfp536 |
| Arid2 | Zkscan7 | Nfxl1 | Zmym2 | Zfp69 | Pf4 | Ywhab | Zfp28 |
| Chaf1b | Fzd8 | Cdca7l | Nr2f2 | Tenm2 | Elk3 | Xpo1 | Fhl5 |
| Ncoa1 | Tgs1 | Ctbp2 | Afap1l2 | Cnot6l | Zfp677 | Dap | Cic |
| Mdk | Col1a1 | Zfp2 | Ptprk | Ret | Zfp266 | Prmt1 | Prmt6 |
| Prmt5 | Arid1a | Cdkn1c | Map3k9 | Basp1 | Zbtb6 | Zbtb5 | Rfx3 |
| Calca | Rhoq | Id2 | Etv6 | Smarce1 | E2f7 | Dnttip2 | Creb1 |
| Ehmt1 | Tcea1 | Zfp229 | Zfp146 | Bcorl1 | Zcchc12 | Smyd2 | Igf2 |
| Cdh13 | Igf1 | Rad21 | Nfya | Pbx1 | Zkscan5 | Mmp14 | Dpf1 |
| Lum | Foxk2 | Zfp318 | Son | Fosl1 | Triap1 | Mafa | Iws1 |
| Wnt11 | Akt1 | Rb1 | Ldb2 | Camta1 | Tgfbr1 | Calr | Dab2 |
| Bcl2l12 | Depdc1a | Hif3a | Zfp551 | Snai2 | Zfp532 | Trp53bp1 | Gtf3a |
| Prdm6 | Txn1 | Prdm8 | Tgif2 | Pnrc2 | Creb3l1 | Mdfic | Ablim1 |
| Ablim3 | Akna | Zfp41 | Zfp46 | Lbx1 | Meis2 | Ndn | Rbm14 |
| Nedd4 | Snapc3 | Snapc1 | Zfp319 | Zfp780b | Med10 | Med11 | Med13 |
| Grem1 | Bcl9l | Ezh2 | Barhl2 | Zfp248 | Hmbox1 | Ralgapa1 | Zfp597 |
| Rrp8 | Prrx1 | Cggbp1 | Chaf1a | Fzd4 | Lcorl | Ywhaq | Ift74 |
| Npas1 | Khdrbs1 | Nr4a3 | Zfp935 | C1d | Med19 | Chd1 | Chek2 |
| Zfp386 | Wwc1 | Tal1 | Hnrnpk | Bcl6b | Hnrnpd | Lims1 | Gtf2a2 |
| Zfp647 | Zfp455 | Fzd3 | Fzd9 | Zfp873 | Myc | Myb | Zfp955b |
| Zfp955a | Osr1 | Osr2 | Nono | Lhx1 | Lhx2 | Zfp52 | Zfp57 |
| Zfp426 | Zfp422 | Tfam | Smad5 | Pa2g4 | Jmy | Zfp334 | Hsf2 |
| Jup | Gpbp1 | Rnf141 | Nfatc4 | Prdm4 | E2f3 | Mlxip | Srsf1 |
| Pspc1 | Rgmb | Meox1 | Vldlr | Sub1 | Phf6 | Zfp592 | Morf4l2 |
| Jak2 | Zic3 | Zic1 | Grm5 | Rbbp8 | Rbbp7 | Cux1 | Ccnc |
| Gm13154 | Med1 | Fzd1 | Armcx3 | Klf7 | Klf5 | Klf3 | Klf8 |
| Snx6 | Picalm | Cdc5l | Atf7 | Ascl1 | Hnrnpab | Zfp652 | Asf1b |
| Hira | Wdr77 | Zfp786 | Srsf10 | Dpy30 | Wwp1 | Zfp81 | Zfp82 |
| Epc1 | Hif1a | Tead2 | Tead1 | Mtdh | Ago1 | Ago2 | Rnf14 |
| Jmjd1c | Zfp326 | Zfp324 | Foxd2 | Foxd1 | Ilf3 | Ilf2 | Dact1 |
| Ddx20 | Crym | Taf10 | Lbh | Kdm6b | Zmynd15 | Zfp619 | Gm6592 |
| Birc5 | Pcgf3 | E2f5 | Khdrbs2 | Bend6 | E2f8 | Smarca1 | Smarca4 |
| Atf2 | Nr4a2 | Kdm5c | Eny2 | Plagl1 | Has3 | Tbl1x | Zfp937 |
| Aebp1 | Aurkb | Tcf12 | Ybx1 | Ybx3 | Tmsb4x | Nr2c2 | Zfp938 |
| Sox11 | Sox12 | Taf1d | Eomes | Zfp78 | Zscan21 | Sertad2 | Trip6 |
| Wwtr1 | Naca | Zfp689 | Mesp1 | Mef2c | Ddx3x | Foxg1 | Mllt11 |
| Zfp945 | Hoxd8 | Mecp2 | Aff4 | Myrfl | Prmt3 | Zfp92 | Zfp128 |
| Tceb1 | Zfp90 | Msx2 | Sox6 | Sox7 | Sox4 | Sox5 | Glis2 |
| Bcl11a | Bcl11b | Csdc2 | Fnip2 | Hinfp | Zmynd11 | Cbx5 | Trip4 |
| Cbx1 | Srebf2 | Cdc73 | Phf20 | 2610305d13rik | Tmf1 | Map3k7 | Bmp2 |
| Ctnnbip1 | 5730507c01rik | Whsc1 | Wac | Hip1 | Zfp397 | Hspa8 | Dnajc17 |
| Myef2 | Mef2a | Hint1 | Rfx7 | Ctnnd1 | Tsc22d2 | Ahr | Taf7 |
| Ash2l | Bclaf1 | Casp8ap2 | Zfp184 | Rbl1 | Ilk | Foxf2 | Foxj3 |
| Hipk2 | Bbs7 | Nrarp | Zfp942 | Dlx1 | Dlx2 | T | Rnf168 |
| Psen1 | Nrip1 | Zfp202 | Lancl2 | Ccna2 | Kdm4b | Kdm4a | Mapk7 |
| Ckap2 | Zfp110 | Rcor3 | Rcor1 | L3mbtl4 | Tnfsf11 | Phf21a | Dnajc2 |
| Smarcc1 | Rps6ka5 | Hells | Heyl | Cdkn1b | Txnip | Scai | Ruvbl2 |
| Nol11 | Osm | Smyd3 | Zbtb41 | Zkscan2 | Zbtb46 | Dr1 | Phtf2 |
| Loxl2 | Zhx3 | Mphosph8 | Mstn | Nab1 | Prkaa2 | Rsf1 | Cd40 |
| Zfp759 | N4bp2l2 | Map2k6 | Maf | Satb2 | Cask | Mlx | Lrpprc |
| Lhx5 | Lhx6 | Lhx9 | Prox1 | Psip1 | Cand1 | Kpna6 | Asf1a |
| Kdm5b | Sp8 | Zfp53 | Usp47 | Sp4 | Csrnp3 | Zfp54 | Zfx |
| Rbmx | Med4 | Dhx36 | Cops2 | Ddx54 | Cops5 | Zfp157 | Hnrnpa2b1 |
| Ctr9 | Mycbp2 | Rbm15b | Skil | Zfp882 | Mina | Jag1 | Zhx1 |
| Ntf3 | Bach1 | Fli1 | Mcrs1 | Ccnt1 | Zkscan17 | Zfp382 | Dach1 |
| Setd1b | Trp73 | Nkap | Zfp583 | Zfp580 | Zfp748 | Pygo1 | Zfp872 |
| Ctnnb1 | Zik1 | Zfp871 | Tenm1 | Taf6l | Zfp366 | Zeb2 | Erbb4 |
| Zfp563 | Ssbp2 | Zfp663 | Asxl3 | Dll1 | Dll4 | Sp1 | Csde1 |
| Dmrt2 | Arf4 | Tbx18 | Zfp174 | Pdpk1 | Kdm2b | Arrb1 | Cdh1 |
| Sfswap | Lmo1 | Tsg101 | Sp5 | Suz12 | Tcerg1 | Brd4 | Tcf4 |
| Gabpb1 | Ascc3 | Ascc2 | Ascc1 | Zfp354c | Nkrf | Pik3r1 | Trim28 |
| Tfap4 | Mycn | Nkx6-1 | Pmf1 | Vps36 | Jazf1 | Cc2d1a | Zfp251 |
| Tet1 | Tet3 | Apbb2 | Dnmt3a | Zfp770 | Mapk15 | Rhoa | Nif3l1 |
| Bzw1 | Pou2f1 | Spdef | Egfr | Pelp1 | Scml4 | Trim27 | Dmrtb1 |
| Phf10 | Foxc1 | Zfp558 | H2afy2 | Aa987161 | Cidea | Eid2 | Eid1 |
| Nfe2l3 | 9030624g23rik | Phf19 | Zmiz1 | Hmgn3 | Hmgn1 | Pcgf2 | Trp53inp1 |
| Sp3 | Nos1 | Smad4 | Smad1 | Smad2 | Sfrp2 | Sfrp1 | Foxq1 |
| Nup62 | Tgfb1i1 | Yy1 | Zfp866 | Elp4 | Elp3 | Zbtb38 | Zbtb33 |
| Brd2 | Brd3 | Olig2 | Pprc1 | Strap | Ets1 | Zfp947 | Zfp946 |
| Zfp944 | Suv420h1 | Suv420h2 | Ssrp1 | Cdk5rap2 | Cdk5rap3 | Thra | Dot1l |
| Vps25 | Pbx3 | Zfp763 | Zfp760 | Cav1 | Btf3 | Zfp493 | Npnt |
| Scmh1 | Foxp2 | Foxp1 | Rbfox2 | Foxp4 | Nr0b1 | Slc40a1 | Wnt7a |
| Zfp697 | Hcfc1 | Irf2bpl | Tnks | Lmo3 | Nipbl | Asxl1 | Atrx |
| Med14 | Bmp7 | Eya2 | Lcor | Trim24 | Zfp449 | Ern2 | Tle3 |
| Tle4 | Pcna | Gatad2b | Pcbd1 | Dnmt1 | Dnmt3b | Ikbkap |
| Pkd1 | Zfhx2 | Scrt1 | Hes3 | Cebpb | Cebpa | Ubp1 | Btg2 |
| Sorbs3 | Tbx6 | Junb | Jund | Mitf | Lime1 | Zfp276 | Zfp513 |
| Foxo4 | Thap11 | Cela1 | Usp2 | Nr1d1 | Nr1d2 | Nkx6-2 | Irf2 |
| Irf1 | Irf5 | Irf8 | Bcas3 | Zbtb17 | Ddit3 | Gtf2h4 | Gtf2f1 |
| Zfp703 | Crebzf | Brms1 | Hdac7 | Litaf | Epas1 | 2410016o06rik | Nacc2 |
| Sec14l2 | Zfp523 | Zfp524 | Ino80b | H2afz | Mypop | Mepce | Tfeb |
| Dkk3 | Rnf6 | Klf16 | Tfe3 | Hace1 | Agap2 | Mapk14 | Mapk11 |
| Mapk12 | Tspyl2 | Gpbp1l1 | Cdk9 | Kcnip3 | Rsc1a1 | Mkx | Zfp827 |
| Zfp14 | Ndnl2 | Wasl | Zfp787 | Mxi1 | Atmin | Med24 | Ttf1 |
| Calcoco1 | Cited4 | Nfic | Cyr61 | Trf | Pkig | Hax1 | Klf10 |
| Klf15 | Bhlhe40 | Bhlhe41 | Fosb | Wnt4 | Fzd2 | Grhl1 | Grhl3 |
| Mkl1 | Mkl2 | Fhl2 | Nr1h2 | Prickle1 | Acvrl1 | Zfp612 | Id4 |
| Nr2f6 | Safb | Supt4a | Dbx2 | Apbb1 | Fgf9 | Fgf1 | Sap30l |
| Zfp692 | Pus1 | Etv3 | Etv5 | Ogt | Gmeb2 | Hlf | Elk1 |
| Zfp664 | Dmrtc1a | Shh | Pbx2 | Stat5b | Thrb | Abra | Zbtb49 |
| Per3 | Per2 | Per1 | Kctd1 | Dbp | Gps2 | Camta2 | Nr1h3 |
| Birc2 | Capn3 | Zfp672 | Pdlim1 | Tesc | Ablim2 | Gata2 | Crem |
| Grin1 | Zfp414 | Rbm10 | Ring1 | Pml | Hdgf | Ezh1 | Aes |
| Esrra | Esrrb | Npas4 | Npas2 | Preb | Sla2 | Ywhah | Hmg20b |
| Taf6 | Safb2 | Snw1 | Zfp36 | Ing2 | Wbscr22 | Pir | Trim11 |
| Nat14 | Egr2 | Egr3 | Egr1 | Egr4 | Rasd1 | Ndufa13 | Nfkbiz |
| Bmyc | Creg1 | Rbbp5 | Wbp2 | Tnip2 | Tnip1 | Ell2 | Pim2 |
| Dvl1 | Skap1 | Dnajb5 | Xrcc5 | Tceal5 | Psmc5 | Sp2 | Hopx |
| Gtf2ird2 | Trerf1 | Zscan25 | Habp4 | Tsc22d3 | Jdp2 | Dyrk1b | Il1a |
| Tcea2 | Cry2 | Pcgf1 | Pcgf5 | Atf5 | Smarca2 | Atf6 | Nr4a1 |
| Erf | Il33 | Tcf15 | Prr13 | Mbd3 | Mbd2 | Arhgef2 | En2 |
| Hexim1 | Sirt2 | Sirt7 | Sox10 | Mical2 | Tfip11 | Sertad1 | Taf11 |
| Zfp688 | Psrc1 | Frzb | Eaf2 | Pou6f1 | Zfp235 | Zfp239 | Zfp93 |
| Nacc1 | Sema4d | Ccdc85b | Sox8 | Fnip1 | Zbtb7a | Fos | Cbx8 |
| Cbx7 | Cbx6 | Cbx4 | Flcn | St18 | Crebl2 | Crebrf | Mdm2 |
| Ctdsp1 | Rorc | Rora | Myrf | S1pr1 | Crtc1 | Crtc2 | Hsf1 |
| Hsf4 | Snf8 | Zfp691 | Fst | Tsc22d4 | Nrip2 | Arid5a | Ddx41 |
| Abcg1 | Zfp112 | Trim66 | Rps6ka4 | Suds3 | Mlxipl | Ell | Prdx5 |
| Nupr1 | Atoh7 | Ccnt2 | Narfl | Nab2 | Wnt10b | Taf8 | Map2k1 |
| Edn1 | Park2 | Prox2 | Cbfa2t3 | Zfp574 | Otud7b | Dapk3 | Nog |
| Csrnp1 | Sp7 | Tbr1 | Med9 | Thrsp | Hexb | Pparg | Ppard |
| Baz1b | Pex14 | Zkscan14 | Zkscan16 | Atp8b1 | Efna1 | Tcf25 | Ldb1 |
| Usf2 | Zfp740 | Zfp367 | Arnt | Pias4 | Pdcd4 | Nlk | Zbtb7b |
| Ccnl2 | Tbl1xr1 | Mapre3 | Pam | Irak1 | Cc2d1b | Trnp1 | Tet2 |
| Batf3 | Tnfrsf4 | Zfp771 | Maf1 | Rhog | Ccnl1 | Nelfe | Mafk |
| E430018j23rik | Hdac11 | Kdm3a | Srf | Bbx | Smad7 | Smad3 | Thap7 |
| Plag1 | Adar | Ovol2 | Ets2 | Mt3 | Pomc | Actr8 | Runx2 |
| Neurog2 | Rreb1 | Ppp3ca | Lmo4 | Tagln3 | Ppm1f | Gclc | Tef |
| Edf1 | Zfpm1 | Rxrb | Ccpg1 | Rxra | E2f6 | Setd3 | Setd7 |
| Zfp846 | Rybp | Bcl9 | Hes6 | 2700060e02rik | Acvr1 | Ccar1 | Zfp712 |
| Mllt1 | Rnasel | Tshz2 | Tshz3 | Sall3 | Sall2 | Drd2 | Runx1t1 |
| Klf11 | Ncl | Gabpa | Mynn | Phb2 | Supt16 | Brip1 | Cdca7 |
| Zfp512 | Jarid2 | Trim33 | Foxo1 | Foxo6 | Atf7ip | Tpr | Kdm1a |
| Kdm1b | Uhrf1 | Top2a | Nme2 | Eapp | Sfpq | Zbtb18 | Bhlhe22 |
| Zfpm2 | Gtf2h5 | Lin54 | Wdr5 | Gtf2f2 | Rbmxl1 | Flna | Dpf3 |
| Hivep2 | Hivep3 | Cebpz | Pou3f3 | Pou3f2 | Pou3f1 | Pou3f4 | Myt1 |
| Hdac2 | Hdac4 | Hdac9 | Notch2 | Notch3 | Phb | Rara | Zfp462 |
| Pogk | Mllt3 | Hmgb2 | Ikzf4 | Hmgb1 | Dmrta1 | Supt20 | Zfp458 |
| Nr2f1 | Fgf10 | Mad2l2 | Bmpr1a | Nhlh2 | L3mbtl1 | L3mbtl3 | Satb1 |
| Ppp2r5d | Ppp2r5b | Foxn3 | Ercc3 | Dmbx1 | Ncoa5 | Zfp282 | Dach2 |
| Rnf2 | Zc3h8 | F2r | Setd2 | Deaf1 | Gsk3b | Spen | Cnot6 |
| Gm13157 | Phf14 | Gtf2e1 | Gtf2e2 | Mospd1 | Hdac10 | Tcfl5 | Cdk2 |
| Tceal7 | Tcf7 | Aatf | Taf9b | Mxd3 | Pid1 | Neurod1 | Neurod6 |
| Ebf2 | Zfp605 | Zfp322a | Zfp72 | Cbfb | Murc | Nfib | Cenpk |
| Usp22 | Xrn2 | Cenpf | Plk1 | Zfml | Ednrb | Rqcd1 | Dvl3 |
| Meaf6 | Cnot1 | Sap130 | Irak1bp1 | Cd36 | Gal | Nfia | Zfp292 |
| Ttf2 | Nfix | Arx | Igsf1 | Snd1 | Pkia | Klf12 | Smarcd3 |
| Arid2 | Nfxl1 | Zmym2 | Pf4 | Zfp28 | Chaf1b | Fzd8 | Cdca7l |
| Nr2f2 | Tenm2 | Elk3 | Xpo1 | Ncoa1 | Tgs1 | Ctbp2 | Afap1l2 |
| Zfp677 | Dap | Cic | Mdk | Col1a1 | Zfp2 | Ptprk | Ret |
| Zfp266 | Prmt1 | Prmt5 | Arid1a | Cdkn1c | Map3k9 | Basp1 | Zbtb6 |
| Zbtb5 | Rfx3 | Calca | Etv6 | Smarce1 | E2f7 | Dnttip2 | Creb1 |
| Ehmt1 | Tcea1 | Zfp229 | Zfp146 | Zcchc12 | Smyd2 | Igf2 | Cdh13 |
| Igf1 | Rad21 | Nfya | Pbx1 | Zkscan5 | Mmp14 | Dpf1 | Lum |
| Foxk2 | Zfp318 | Mafa | Iws1 | Rb1 | Ldb2 | Camta1 | Calr |
| Dab2 | Hif3a | Zfp551 | Snai2 | Trp53bp1 | Gtf3a | Txn1 | Tgif2 |
| Pnrc2 | Creb3l1 | Mdfic | Ablim1 | Ablim3 | Zfp41 | Zfp46 | Meis2 |
| Ndn | Rbm14 | Nedd4 | Zfp319 | Med11 | Med13 | Grem1 | Bcl9l |
| Ezh2 | Barhl2 | Hmbox1 | Ralgapa1 | Zfp597 | Prrx1 | Cggbp1 | Fzd4 |
| Lcorl | Ywhaq | Khdrbs1 | Nr4a3 | Zfp935 | C1d | Med19 | Wwc1 |
| Hnrnpk | Bcl6b | Hnrnpd | Lims1 | Gtf2a2 | Zfp647 | Zfp455 | Fzd3 |
| Fzd9 | Myc | Myb | Zfp955b | Zfp955a | Osr1 | Nono | Lhx1 |
| Lhx2 | Zfp52 | Zfp57 | Zfp426 | Zfp422 | Smad5 | Pa2g4 | Jmy |
| Hsf2 | Jup | Gpbp1 | Rnf141 | Nfatc4 | Prdm4 | Mlxip | Srsf1 |
| Pspc1 | Rgmb | Meox1 | Vldlr | Phf6 | Jak2 | Zic3 | Zic1 |
| Grm5 | Rbbp8 | Rbbp7 | Ccnc | Med1 | Armcx3 | Klf7 | Klf3 |
| Klf8 | Snx6 | Picalm | Cdc5l | Ascl1 | Hnrnpab | Zfp652 | Asf1b |
| Hira | Wdr77 | Zfp786 | Srsf10 | Dpy30 | Wwp1 | Zfp81 | Epc1 |
| Hif1a | Tead2 | Tead1 | Mtdh | Ago1 | Ago2 | Rnf14 | Jmjd1c |
| Zfp326 | Zfp324 | Foxd1 | Ilf3 | Ilf2 | Dact1 | Crym | Lbh |
| Kdm6b | Zmynd15 | Zfp619 | Gm6592 | Birc5 | Pcgf3 | Khdrbs2 | Bend6 |
| E2f8 | Smarca1 | Smarca4 | Kdm5c | Eny2 | Has3 | Tbl1x | Zfp937 |
| Aebp1 | Aurkb | Tcf12 | Ybx1 | Ybx3 | Tmsb4x | Nr2c2 | Zfp938 |
| Sox11 | Sox12 | Taf1d | Zscan21 | Wwtr1 | Zfp689 | Mesp1 | Mef2c |
| Ddx3x | Foxg1 | Mllt11 | Zfp945 | Mecp2 | Aff4 | Zfp92 | Zfp128 |
| Tceb1 | Zfp90 | Sox6 | Sox7 | Sox4 | Sox5 | Glis2 | Bcl11a |
| Bcl11b | Csdc2 | Fnip2 | Hinfp | Cbx5 | Cbx1 | Cdc73 | Phf20 |
| 2610305d13rik | Tmf1 | Ctnnbip1 | Whsc1 | Wac | Hip1 | Zfp397 | Dnajc17 |
| Myef2 | Mef2a | Rfx7 | Ctnnd1 | Ahr | Taf7 | Ash2l | Bclaf1 |
| Casp8ap2 | Zfp184 | Rbl1 | Ilk | Foxf2 | Nrarp | Dlx1 | Rnf168 |
| Nrip1 | Zfp202 | Lancl2 | Ccna2 | Kdm4b | Kdm4a | Mapk7 | Ckap2 |
| Zfp110 | Rcor3 | Rcor1 | L3mbtl4 | Dnajc2 | Smarcc1 | Rps6ka5 | Hells |
| Heyl | Cdkn1b | Txnip | Scai | Nol11 | Smyd3 | Zbtb41 | Zbtb46 |
| Dr1 | Phtf2 | Loxl2 | Zhx3 | Mphosph8 | Mstn | Nab1 | Prkaa2 |
| Rsf1 | Cd40 | Map2k6 | Maf | Satb2 | Cask | Lrpprc | Lhx5 |
| Lhx6 | Prox1 | Psip1 | Cand1 | Asf1a | Kdm5b | Zfp53 | Usp47 |
| Csrnp3 | Zfp54 | Zfx | Rbmx | Med4 | Dhx36 | Cops5 | Zfp157 |
| Hnrnpa2b1 | Ctr9 | Rbm15b | Skil | Zfp882 | Jag1 | Zhx1 | Ntf3 |
| Bach1 | Fli1 | Mcrs1 | Ccnt1 | Zfp382 | Dach1 | Setd1b | Trp73 |
| Nkap | Zfp583 | Zfp580 | Zfp748 | Pygo1 | Ctnnb1 | Zik1 | Tenm1 |
| Zeb2 | Erbb4 | Zfp563 | Ssbp2 | Zfp663 | Asxl3 | Dll1 | Dll4 |
| Dmrt2 | Arf4 | Tbx18 | Zfp174 | Pdpk1 | Kdm2b | Cdh1 | Sfswap |
| Lmo1 | Sp5 | Suz12 | Brd4 | Tcf4 | Ascc3 | Ascc1 | Zfp354c |
| Nkrf | Pik3r1 | Trim28 | Tfap4 | Mycn | Vps36 | Jazf1 | Cc2d1a |
| Zfp251 | Tet1 | Tet3 | Apbb2 | Dnmt3a | Zfp770 | Mapk15 | Rhoa |
| Bzw1 | Pou2f1 | Spdef | Egfr | Pelp1 | Scml4 | Trim27 | Dmrtb1 |
| Phf10 | Foxc1 | Zfp558 | H2afy2 | Cidea | Eid2 | Eid1 | Nfe2l3 |
| 9030624g23rik | Phf19 | Zmiz1 | Hmgn3 | Pcgf2 | Trp53inp1 | Sp3 | Nos1 |
| Smad4 | Smad1 | Smad2 | Sfrp2 | Sfrp1 | Nup62 | Tgfb1i1 | Yy1 |
| Zfp866 | Elp4 | Elp3 | Zbtb33 | Brd2 | Brd3 | Pprc1 | Strap |
| Ets1 | Zfp946 | Zfp944 | Suv420h1 | Suv420h2 | Cdk5rap2 | Thra | Dot1l |
| Pbx3 | Zfp763 | Zfp760 | Cav1 | Btf3 | Npnt | Scmh1 | Foxp2 |
| Foxp1 | Rbfox2 | Foxp4 | Nr0b1 | Slc40a1 | Wnt7a | Zfp697 | Hcfc1 |
| Irf2bpl | Tnks | Lmo3 | Nipbl | Asxl1 | Atrx | Med14 | Bmp7 |
| Eya2 | Lcor | Trim24 | Tle3 | Tle4 | Gatad2b | Pcbd1 | Dnmt1 |
| Dnmt3b | Ikbkap |
| Pkd1 | Zfhx2 | Cebpa | Btg2 | Sorbs3 | Junb | Jund | Mitf |
| Zfp513 | Foxo4 | Cela1 | Usp2 | Nr1d1 | Nr1d2 | Nkx6-2 | Irf2 |
| Irf1 | Irf5 | Irf8 | Bcas3 | Zbtb17 | Ddit3 | Zfp703 | Brms1 |
| Litaf | Epas1 | Sec14l2 | Zfp523 | Zfp524 | Mypop | Tfeb | Dkk3 |
| Rnf6 | Klf16 | Hace1 | Mapk14 | Mapk11 | Mapk12 | Tspyl2 | Cdk9 |
| Kcnip3 | Mkx | Zfp827 | Ndnl2 | Wasl | Mxi1 | Atmin | Med24 |
| Calcoco1 | Cited4 | Nfic | Cyr61 | Trf | Pkig | Hax1 | Klf10 |
| Klf15 | Bhlhe40 | Bhlhe41 | Fosb | Wnt4 | Grhl1 | Mkl2 | Fhl2 |
| Nr1h2 | Prickle1 | Zfp612 | Id4 | Nr2f6 | Supt4a | Fgf1 | Etv3 |
| Etv5 | Hlf | Elk1 | Zfp664 | Dmrtc1a | Pbx2 | Stat5b | Per3 |
| Per2 | Per1 | Kctd1 | Dbp | Camta2 | Birc2 | Capn3 | Pdlim1 |
| Tesc | Ablim2 | Crem | Grin1 | Rbm10 | Ring1 | Ezh1 | Esrra |
| Npas4 | Preb | Sla2 | Hmg20b | Safb2 | Snw1 | Zfp36 | Ing2 |
| Wbscr22 | Pir | Trim11 | Nat14 | Egr2 | Egr3 | Egr1 | Egr4 |
| Rasd1 | Ndufa13 | Bmyc | Creg1 | Wbp2 | Tnip2 | Tnip1 | Ell2 |
| Pim2 | Dvl1 | Skap1 | Dnajb5 | Tceal5 | Psmc5 | Sp2 | Hopx |
| Gtf2ird2 | Trerf1 | Zscan25 | Habp4 | Tsc22d3 | Jdp2 | Dyrk1b | Tcea2 |
| Cry2 | Pcgf1 | Pcgf5 | Atf5 | Smarca2 | Atf6 | Nr4a1 | Erf |
| Il33 | Prr13 | Mbd3 | Mbd2 | Arhgef2 | En2 | Hexim1 | Sirt2 |
| Sirt7 | Sox10 | Mical2 | Tfip11 | Sertad1 | Taf11 | Zfp688 | Psrc1 |
| Frzb | Pou6f1 | Zfp235 | Zfp239 | Sema4d | Ccdc85b | Sox8 | Zbtb7a |
| Fos | Cbx8 | Cbx7 | Cbx6 | Flcn | St18 | Crebl2 | Crebrf |
| Mdm2 | Rora | Myrf | S1pr1 | Crtc2 | Hsf1 | Hsf4 | Zfp691 |
| Fst | Tsc22d4 | Nrip2 | Arid5a | Abcg1 | Trim66 | Rps6ka4 | Suds3 |
| Mlxipl | Ell | Nupr1 | Atoh7 | Ccnt2 | Narfl | Nab2 | Wnt10b |
| Taf8 | Map2k1 | Edn1 | Park2 | Zfp574 | Otud7b | Dapk3 | Csrnp1 |
| Sp7 | Tbr1 | Med9 | Thrsp | Hexb | Pparg | Ppard | Baz1b |
| Pex14 | Zkscan14 | Zkscan16 | Efna1 | Ldb1 | Usf2 | Zfp740 | Zfp367 |
| Arnt | Nlk | Zbtb7b | Mapre3 | Pam | Irak1 | Cc2d1b | Trnp1 |
| Batf3 | Zfp771 | Maf1 | Rhog | E430018j23rik | Hdac11 | Kdm3a | Srf |
| Smad7 | Smad3 | Thap7 | Plag1 | Adar | Ovol2 | Ets2 | Mt3 |
| Actr8 | Runx2 | Neurog2 | Rreb1 | Ppp3ca | Lmo4 | Tagln3 | Ppm1f |
| Gclc | Tef | Zfpm1 | Rxra | Setd3 | Setd7 |
| Zfp846 | Rybp | Bcl9 | Hes6 | 2700060e02rik | Ccar1 | Zfp712 | Mllt1 |
| Tshz2 | Tshz3 | Sall2 | Drd2 | Runx1t1 | Klf11 | Ncl | Supt16 |
| Brip1 | Cdca7 | Zfp512 | Trim33 | Foxo1 | Foxo6 | Atf7ip | Tpr |
| Kdm1a | Kdm1b | Uhrf1 | Top2a | Nme2 | Sfpq | Zbtb18 | Bhlhe22 |
| Zfpm2 | Gtf2h5 | Wdr5 | Gtf2f2 | Rbmxl1 | Flna | Hivep2 | Hivep3 |
| Cebpz | Pou3f3 | Pou3f2 | Pou3f1 | Myt1 | Hdac2 | Notch3 | Zfp462 |
| Pogk | Mllt3 | Hmgb2 | Ikzf4 | Hmgb1 | Dmrta1 | Supt20 | Zfp458 |
| Fgf10 | Bmpr1a | L3mbtl1 | L3mbtl3 | Satb1 | Ppp2r5d | Ppp2r5b | Foxn3 |
| Ncoa5 | Zfp282 | Dach2 | Rnf2 | F2r | Setd2 | Deaf1 | Gsk3b |
| Cnot6 | Phf14 | Gtf2e1 | Gtf2e2 | Mospd1 | Tcfl5 | Tceal7 | Tcf7 |
| Mxd3 | Pid1 | Neurod1 | Neurod6 | Zfp605 | Zfp322a | Zfp72 | Cbfb |
| Nfib | Cenpk | Usp22 | Xrn2 | Cenpf | Zfml | Ednrb | Rqcd1 |
| Meaf6 | Cnot1 | Cd36 | Gal | Nfia | Ttf2 | Nfix | Igsf1 |
| Pkia | Klf12 | Arid2 | Pf4 | Chaf1b | Fzd8 | Nr2f2 | Tenm2 |
| Xpo1 | Tgs1 | Ctbp2 | Afap1l2 | Zfp677 | Cic | Mdk | Col1a1 |
| Ptprk | Ret | Zfp266 | Prmt1 | Prmt5 | Basp1 | Zbtb6 | Zbtb5 |
| Rfx3 | Calca | Etv6 | Smarce1 | E2f7 | Dnttip2 | Creb1 | Ehmt1 |
| Tcea1 | Zfp229 | Zfp146 | Zcchc12 | Smyd2 | Igf2 | Cdh13 | Igf1 |
| Nfya | Pbx1 | Mmp14 | Dpf1 | Lum | Foxk2 | Zfp318 | Rb1 |
| Ldb2 | Camta1 | Dab2 | Hif3a | Zfp551 | Trp53bp1 | Gtf3a | Txn1 |
| Creb3l1 | Ablim1 | Ablim3 | Zfp41 | Zfp46 | Ndn | Nedd4 | Zfp319 |
| Med11 | Med13 | Bcl9l | Ezh2 | Hmbox1 | Ralgapa1 | Zfp597 | Cggbp1 |
| Lcorl | Ywhaq | Khdrbs1 | Nr4a3 | Zfp935 | C1d | Med19 | Wwc1 |
| Bcl6b | Hnrnpd | Gtf2a2 | Zfp647 | Fzd3 | Fzd9 | Myc | Zfp955b |
| Zfp955a | Nono | Lhx2 | Zfp57 | Zfp426 | Zfp422 | Smad5 | Pa2g4 |
| Jmy | Hsf2 | Jup | Rnf141 | Nfatc4 | Mlxip | Srsf1 | Pspc1 |
| Rgmb | Meox1 | Vldlr | Phf6 | Zic3 | Zic1 | Grm5 | Armcx3 |
| Klf7 | Klf3 | Klf8 | Snx6 | Picalm | Ascl1 | Hnrnpab | Wdr77 |
| Zfp786 | Srsf10 | Wwp1 | Zfp81 | Epc1 | Hif1a | Tead2 | Tead1 |
| Mtdh | Ago1 | Ago2 | Jmjd1c | Zfp326 | Zfp324 | Ilf2 | Dact1 |
| Lbh | Kdm6b | Birc5 | Pcgf3 | Khdrbs2 | Bend6 | Smarca1 | Has3 |
| Tbl1x | Aurkb | Ybx1 | Ybx3 | Tmsb4x | Nr2c2 | Sox11 | Sox12 |
| Taf1d | Wwtr1 | Zfp689 | Mesp1 | Mef2c | Mllt11 | Zfp945 | Mecp2 |
| Aff4 | Zfp92 | Zfp128 | Tceb1 | Zfp90 | Sox6 | Sox7 | Sox4 |
| Sox5 | Glis2 | Bcl11a | Bcl11b | Csdc2 | Fnip2 | Cbx5 | Cbx1 |
| Ctnnbip1 | Whsc1 | Wac | Hip1 | Zfp397 | Myef2 | Mef2a | Rfx7 |
| Ctnnd1 | Ash2l | Rbl1 | Ilk | Foxf2 | Nrarp | Dlx1 | Rnf168 |
| Nrip1 | Lancl2 | Ccna2 | Kdm4b | Kdm4a | Mapk7 | Ckap2 | Zfp110 |
| Rcor3 | Rcor1 | L3mbtl4 | Dnajc2 | Smarcc1 | Hells | Cdkn1b | Txnip |
| Scai | Smyd3 | Zbtb41 | Dr1 | Phtf2 | Loxl2 | Zhx3 | Mstn |
| Nab1 | Prkaa2 | Map2k6 | Maf | Satb2 | Cask | Lrpprc | Lhx6 |
| Psip1 | Cand1 | Kdm5b | Usp47 | Csrnp3 | Rbmx | Dhx36 | Cops5 |
| Hnrnpa2b1 | Ctr9 | Rbm15b | Skil | Zhx1 | Bach1 | Mcrs1 | Ccnt1 |
| Dach1 | Trp73 | Nkap | Zfp583 | Zfp580 | Zfp748 | Zik1 | Tenm1 |
| Zeb2 | Erbb4 | Zfp563 | Ssbp2 | Asxl3 | Dll1 | Dll4 | Dmrt2 |
| Arf4 | Tbx18 | Pdpk1 | Kdm2b | Sp5 | Tcf4 | Ascc3 | Ascc1 |
| Zfp354c | Nkrf | Pik3r1 | Mycn | Vps36 | Jazf1 | Cc2d1a | Zfp251 |
| Tet1 | Tet3 | Apbb2 | Dnmt3a | Zfp770 | Mapk15 | Bzw1 | Egfr |
| Scml4 | Dmrtb1 | Phf10 | Zfp558 | H2afy2 | Eid1 | Nfe2l3 | 9030624g23rik |
| Zmiz1 | Nos1 | Smad4 | Smad1 | Smad2 | Sfrp2 | Sfrp1 | Nup62 |
| Tgfb1i1 | Yy1 | Elp4 | Elp3 | Zbtb33 | Brd2 | Brd3 | Pprc1 |
| Ets1 | Zfp946 | Suv420h1 | Suv420h2 | Dot1l | Pbx3 | Zfp763 | Zfp760 |
| Cav1 | Btf3 | Npnt | Foxp1 | Rbfox2 | Slc40a1 | Tnks | Lmo3 |
| Nipbl | Atrx | Eya2 | Trim24 | Tle3 | Tle4 | Gatad2b | Pcbd1 |
| Ikbkap |
| Ascl3 | Zfp652 | Tshz2 | Zfp69 | Srrt | Gm6592 | Dot1l | Flna |
| Hivep3 | Per1 | Hdac7 | Zfp455 | L3mbtl1 | Cc2d1b | Hipk2 | Spen |
| Hdac10 | Hif3a | Snapc4 | Med12 | Zfp169 | Hmbox1 | Zbtb16 | Zfp692 |
| Acvr2b | Nrip2 | Trip6 | Msx1 | Txnip | Pitx1 | Batf2 | Zhx3 |
| Nos1 | Tle2 | Id4 | Lmx1b | Ar | Ebf4 | Jak3 |
| Hif3a | Snapc4 | Tshz2 | Pitx1 | Srrt | Ebf4 | Batf2 | Tle2 |
| Gm6592 | L3mbtl1 | Cc2d1b |
| Tle2 | Gm6592 |
| Med13l | Zfp846 | Tgs1 | Drd3 | Csrnp3 | Zfp57 | Ccar1 | Runx1t1 |
| Klf12 | Pnn | Zkscan16 | Ago1 | Ago2 | Scai | Supt16 | Zfp445 |
| Hlf | Zbed6 | Ppp1r12a | Atf7ip | Pcgf5 | Foxo3 | Ppargc1b | Atf7 |
| Elk4 | Inpp5k | Gabpb2 | Tbl1x | Asxl2 | Mlxip | Flna | Hivep3 |
| Nr2c2 | Per2 | Pou3f2 | Taf1d | Pik3r1 | Supt20 | Ankrd49 | Arid4a |
| Arid4b | Son | Banp | N4bp2l2 | Ash1l | L3mbtl1 | Satb2 | Foxn3 |
| Ncoa7 | Pnrc2 | Rb1cc1 | Erbb4 | Hipk2 | Mafg | Hinfp | Tcerg1 |
| Spen | Zfp354c | Gcfc2 | Med12 | Rsc1a1 | Prkdc | Hmbox1 | Nipbl |
| Bdp1 | Il4 | Zbtb33 | Chd2 | Ahctf1 | Nrip1 | Cnot1 | Arid5b |
| Atxn7 | Rcor1 | Txlng | Npnt | Jmy | Rps6ka5 | Mdm4 | Mysm1 |
| Zkscan2 | Tnks | Insr | Atrx | Lcor | Rsf1 | Nck1 | Tenm1 |
| Map2k6 | Foxp3 | Ar | Setd7 |
| Med13l | Zfp846 | Nrip1 | Erbb4 | Hipk2 | Tbl1x | Npnt | Hivep3 |
| Taf1d | Rsc1a1 | Zkscan2 | Hmbox1 | Zfp445 | Hlf | Supt20 | Il4 |
| Zbed6 | Map2k6 | Atf7ip | Son |
| Taf1d | Rsc1a1 | Zkscan2 | Zbed6 | Hivep3 |
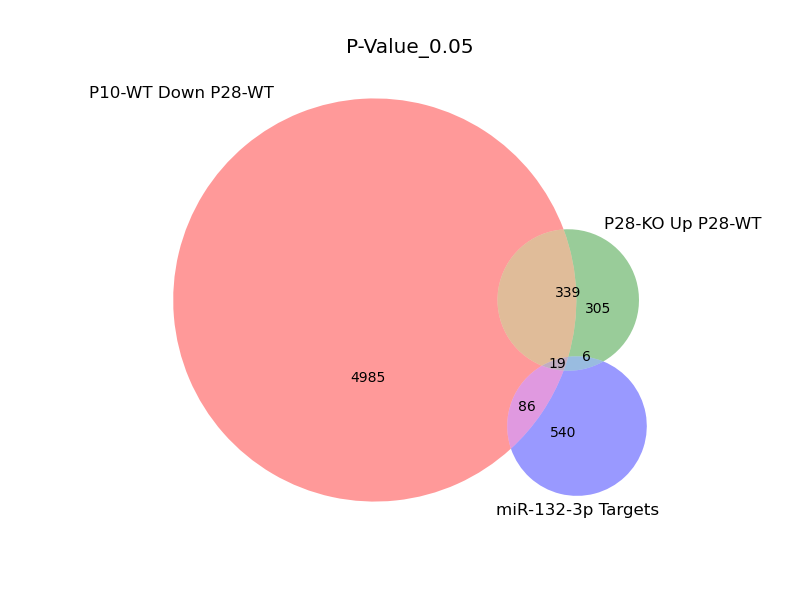 |
 |
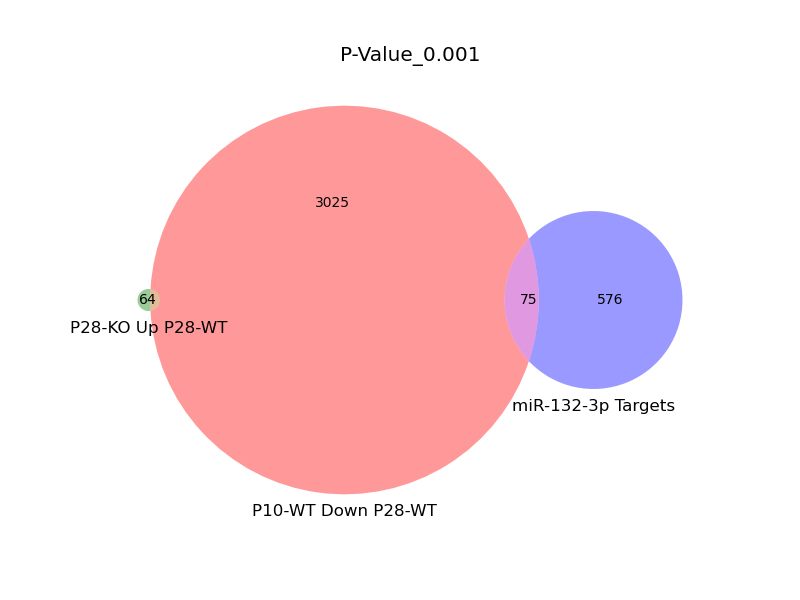 |